Mutations in a number of genes are now known to cause susceptibility to breast cancer. In the context of high-risk families, the most important genes are BRCA1 and BRCA2. Inherited mutations in BRCA1/2 may be necessary to explain the Mendelian pattern of breast cancer in some families but are not sufficient to completely describe interindividual variability in cancer risk. In this work we tried to identify possible modifier genes in a group of hereditary, sporadic breast cancers and controls by gene expression profiling.
We show that normal breast tissues collected from hereditary breast cancer patients and from healthy BRCA1/2 gene mutation carriers reveal a significant different molecular profile compared to the normal tissue of sporadic breast cancer patients and healthy BRCA1/2 gene mutation non-carriers. The present study demonstrates that TC1 and SYNGR2 genes are statistically overexpressed not only in tumoral tissues but also in the normal tissues of hereditary breast cancer patients and in BRCA1/2 healthy carriers compared to sporadic breast cancer patients or healthy controls. Silencing of SYNGR2 and TC1 was performed on MCF7 breast cancer cell line to better understand their involvement in breast cancer. Our results suggest that normal mammary tissue of BRCA1/2 gene carriers could be another source for the discovery of other genetic determinants that favor hereditary cancer onset.
Breast cancer is the most common cancer in women worldwide, with nearly 1.7 million new cases diagnosed in 2012 [1]. Hereditary breast cancer (HBC) constitute 5% to 10% of breast cancer cases overall [2]. Nowadays it is well known that mutations in various genes can confer susceptibility to the development of breast cancer. In the context of high-risk families, the most important genes are BRCA1 and BRCA2. The alteration of BRCA1/2 genes predisposes to a high lifetime risk of cancer but also influences the biology of the tumor in a characteristic way. These patients exhibit an inherent molecular stigma, which clearly distinguishes them from sporadic breast cancer (SBC) patients. However inherited mutations in BRCA1/2 may explain the Mendelian model of breast cancer in some families, but they are not sufficient to fully describe the inter-individual variability in cancer risk. Several studies suggest that additional modifying factors could influence cancer penetrance among BRCA1/2 mutation carriers [3]. These could be both lifestyle, hormone and genetic factors. In this regard in 2007, the Consortium of Investigators of Modifiers of BRCA1 and BRCA2 (CIMBA) has been established, to identify modifier genes [4].
The aim of this work was to identify possible modifier genes in the normal mammary tissues of a series of HBCs compared to SBCs and controls by performing gene expression profiling.
In this regard, we used a specific cancer gene panel previously proven to discriminate benign and malignant tumors [5,6] and which included several genes already involved in breast cancer. We focused then our attention on TC1 and SYNGR2 genes as possible modifier genes. Furthermore, TC1 and SYNGR2 silencing was performed in MCF7 breast cancer cell line to investigate their involvement in neoplastic progression.
Samples
As shown in table 1 our case study was composed of 42 tissue samples: normal mammary tissues (NMT) from 9 HBC BRCA1/2 carrier patients (NMT-HBC BRCA1/2), normal mammary tissues from 4 healthy (H) BRCA1/2 carrier individuals (NMT-H BRCA1/2), normal mammary tissues from 8 SBC patients (NMT-SBC), normal mammary tissues from 4 healthy BRCA1/2 non-carrier individuals (NMT-H), tumor mammary tissues (TMT) from 9 HBC BRCA1/2 carrier patients (TMT-HBC BRCA1/2) and 8 tumor mammary tissues from SBC (TMT-SBC). HBC BRCA1/2 carrier patients and healthy BRCA1/2 carrier individuals were collected from the Department of Cytogenetics and Molecular Genetic Unit of the University Hospital of Pisa, Italy. The SBC patients and healthy BRCA1/2 non-carrier individuals, which underwent reductive mastectomy, were selected from the archives of the Anatomy Pathology Institute of the University Hospital of Pisa, Italy. For each cancer patient we selected the tumoral and normal mammary tissue. For each tissue block 2 sections of 10 μm thickness were cut using a new microtome blade and collected in a 1.5 ml tube.
Table 1. Sample list. List of all the samples used for the gene expression analyses
Samples |
Numbers |
Abbreviations |
Normal Mammary Tissues from HBC BRCA1/2 carrier patients |
9 |
NMT-HBC BRCA1/2 |
Normal Mammary Tissues from Healthy BRCA1/2 carrier individuals |
4 |
NMT-H BRCA1/2 |
Normal Mammary Tissues from SBC |
8 |
NMT-SBC |
Normal Mammary Tissues from Healthy BRCA1/2 non-carrier individuals |
4 |
NMT-H |
Tumor Mammary Tissues from BRCA1/2 carrier HBC patients |
9 |
TMT-HBC BRCA1/2 |
Tumor Mammary Tissues from SBC |
8 |
TMT-SBC |
All samples |
42 |
|
Ethical board
The study was approved by the Ethics Committee of University Hospital of Pisa and signed informed consent was obtained from each of the subjects. All methods were performed in accordance with approved guidelines.
Gene panel
We analysed our samples with a gene panel already used in our previous papers [5,6] able to discriminate benign and malignant tumors in thyroid carcinoma patients. The gene panel was composed of the following genes: SYNGR2, TC1, LSM7, CDH1, CKIT, DOC1, KLF4, DDI2, Hs296031.
RNA isolation
RNA extraction was performed using the RNeasy Mini kit (Qiagen, Venlo, Netherlands) according to manufacturer’s protocol. RNA extracted was treated with DNase Ι recombinant, RNase-free (Roche, Basel, Switzerland).
Real time PCR
RNA was reverse transcribed in a final volume of 20 μl, containing 5X RT buffer, 10 mM dNTPs, 50 ng/μl Random Primers, 0.1 M DTT, 40 U/μl RNaseOUT, 50 μM oligo(dT), DEPC-Treated Water, 15 U/μl Cloned AMV reverse transcriptase (Invitrogen, Carlsbad, CA). The level of SYNGR2, TC1, LSM7, CDH1, CKIT, DOC, KLF4, DDI2, Hs296031 mRNA and miR128 and miR210 was analyzed by quantitative Real Time PCR (qPCR) on the Rotor Gene 6000 real time rotary analyzer (Corbett Life Science, Sidney, Australia) following the manufacturer’s instructions. Endogenous reference gene (B2M, beta 2 microglobulin) was used to normalize each gene expression level. PCR was performed in 25 μl final volume, containing 5 μl of cDNA, 12.5 μl of MESA GREEN qPCR MasterMix Plus (Eurogentec, San Diego, CA, USA), 300 nM of each primer (Invitrogen, Carlsbad, CA, USA) with the following cycling conditions: initial denaturation 95˚C for 5 min; 40 cycles at 95˚C for 15 sec and 58˚C for 40 sec and 72˚C for 40 sec; final step 25˚C for 1 min. Each samples was performed in triplicate. The primers for all the gene are shown in table 2.
Table 2. Primers sequences. List of the primers sequences (forward and reverse), used for gene expression analyses
Gene |
Primer Sequence |
CKIT_Forward |
5’- GCACCTGCTGCTGAAATGTATGACATAAT - 3’ |
CKIT_Reverse |
5’- TTTGCTAAGTTGGAGTAAATATGATTGG - 3’ |
SYNGR2_Forward |
5’- ATCTTCTCCTGGGGTGTGCT - 3’ |
SYNGR2_Reverse |
5’- AGGGTGGCTGTTGGTAGTTG - 3’ |
Hs.296031_Forward |
5’- TGCCAAGGAGCTTTATAGAA - 3’ |
Hs.296031_Reverse |
5’- ATGACGGCATGTACCAACCA - 3’ |
DDI2_Forward |
5’- TGCAGTTCCCAAACTTACCC- 3’ |
DDI2_Reverse |
5’- CAGCAACATATCTCGGAGCA- 3’ |
CDH1_Forward |
5’- GCATTGCCACATACACTCTC- 3’ |
CDH1_Reverse |
5’- AGCACCTTCCATGACAGAC- 3’ |
LSM7_Forward |
5’-GACGATCCGGGTAAAGTTCCA - 3’ |
LSM7_Reverse |
5’- AGGTTGAGGAGTGGGTCGAA - 3’ |
TC1_Forward |
5’- AAATCTTCTGACTAATGCTAAAACG - 3’ |
TC1_Reverse |
5’- TTATTGTTGCATGACATTTGC - 3’ |
KLF4_Forward |
5′-TTGGCTTCGTTTCTTCTCTTCGTTGACTTT-3′ |
KLF4_Reverse |
5′-CGGAGGATGGGTCAGCGAATT-3′ |
DOC_Forward |
5'-AGTCCATACTGATATTTTTGCAAG GAA-3' |
DOC_Reverse |
5'-CCCAAAGTACGAGTTCAGTCAGTCT-3' |
B2M_Forward |
5’- CATTCCTGAAGCTGACAGCATTC - 3’ |
B2M_Reverse |
5’- TGCTGGATGACGTGAGTAAACC - 3’ |
Cell culture
The human cancer cell line MCF7 (ATCC, LGC Standards, Manassas, VA, USA) was cultured in high glucose DMEM (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% FBS (Thermo Fisher Scientific, Waltham, MA, USA), 100 U/ml penicillin (Thermo Fisher Scientific, Waltham, MA, USA) and 100 μg/ml streptomycin (Thermo Fisher Scientific, Waltham, MA, USA) at 37°C in an atmosphere of 5% CO2.
TC1 and SYNGR2 silencing
Sure Silencing shRNA Plasmids (Qiagen, Venlo, Netherlands) were used to silence TC1 and SYNGR2 mRNA in MCF7 cells following the manufacturer’s instructions. We generated MCF7-ShTC1 and MCF7-ShSYNGR2 cells and as control MCF7-TC1-Ctrl and MCF7-SYNGR2-Ctrl treated with the negative plasmid found in the kit.
First of all we generated the dose response curve to determine the minimum antibiotic concentration (neomycin) necessary to kill the untransfected cells in order to create stable cells. The antibiotic concentration was 1.6 mg/ml.
Real Time PCR
Total RNA was extracted from the transfected cells using Maxwell® 16 LEV SimplyRNA Purification Kit (Promega, Madison, WI, USA) following exactly the manufacturer’s protocol. Reverse transcription was performed with Nanoscript 2 Reverse Transcription kit (PrimerDesign, Chandler’s Ford, United Kingdom) as instructed. Real time PCR was performed using Sso Advanced Universal SYBR Green Supermix (Biorad, Hercules, CA, USA) in a final volume of 20 μl with 10 μl of Sso Advanced Universal SYBR Green Supermix and 20X specific primer assay. The primers used were the C8orf4 (TC1) PrimePCR™ SYBR® Green Assay and the SYNGR2 PrimePCR™ SYBR® Green Assay (Biorad, Hercules, CA, USA). Endogenous reference gene (ACTB, beta actin) was used to normalize each gene expression level.
Western blot
Cells were digested with trypsin and centrifuged. The cell pellet was washed twice with PBS. Next, the cells were treated with RIPA buffer (150 mM NaCl, 1.0% IGEPAL® CA-630, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris, pH 8.0) (Sigma Aldrich, St. Louis, MO, USA) and Protease/Phosphatase Inhibitor Cocktail (100X) (Cell Signaling Technologies, Danvers, MA, USA) on ice for 30 min and centrifuged at 12,000 rpm for 15 min. Cell lysates (30 mg/well) were subjected to SDS-PAGE and transferred to nitrocellulose filter membranes. The membranes were incubated with primary antibodies: TC1 (C-13) (sc-98165, polyclonal) (Santa Cruz Biotechnology, Dallas, TE, USA), anti-SYNGR2 (HPA014742, polyclonal) (Sigma Aldrich, St. Louis, MO, USA) and anti-TUBB2B (SAB1402092, polyclonal) (Sigma Aldrich, St. Louis, MO, USA) as control. Secondary antibodies conjugated with horseradish peroxidase were subsequently used. Signals were detected using Clarity Western ECL (Biorad, Hercules, CA, USA) and Chemidoc Imager (Biorad, Hercules, CA, USA). The results were analyzed using Image J Tool.
Immunocytochemistry
40,000 cells of all kinds were grown on a 2-well tissue culture chambers on glass slide (Sarstedt, Numbrecht, Germany). The day after the glass slides were fixed with 3.7% formaldehyde for 10 minutes and washed three times in PBS. Cells were permeabilized with 0.1% Triton X-100 in and washed three times in PBS. Immunocytochemistry was performed using the Mouse specific HRP/DAB (ABC) Detection IHC Kit (Abcam, Cambridge, UK) according to manufacturer’s protocol. The antigen unmasking was achieved with MS-unmasker solution (DIAPATH, Martinengo, BG, Italy) in microwave. Primary antibodies, TC1 (C-13) (Santa Cruz Biotechnology, Dallas, TE, USA) and anti-SYNGR2 (Sigma Aldrich, St. Louis, MO, USA) was used at 1:200 dilution for 1h at room temperature. Slides were developed with diaminobenzidine chromogen (DAB) (DAKO, Glostrup, DK) and counterstained with hematoxylin. Negative controls included the omission of the primary antibody. Slides were analyzed using the inverted microscope CARL ZEISS Axio Observer Z1FLMot, and images were taken with CARL ZEISS AXIOCAM Icc1 camer. The percentage of staining was evaluated with ImageJ software analyzing 30 different fields per slide.
WST1 viability assay
A total of 4,000 cells per well of each type (MCF7-ShTC1, MCF7-ShSYNGR2, MCF7-TC1-Ctrl and MCF7-SYNGR2-Ctrl) were seeded in a 96-well plate format, after 48h from transfection. After 24h the WST1 reagent (Clontech Laboratories, Mountain View, CA, USA) was added and incubated for a further 2h before reading the plate. Each assay was conducted in sets of seven. The quantity of formazan dye is directly related to the number of metabolically active cells, and was quantified by measuring the absorbance at 450 nm in a multiwell plate reader (Tecan, Mannedorf, Switzerland).
Wound healing assay
30,000 cells of each kind were seeded in each insert of a dish with a culture-insert 2 well (IBIDI, Martinsried, Germany). When the cells reached 90% confluence in the dish, the insert was removed. The debris was removed by washing with serum free medium. After 24 h of incubation, the cells which migrated into the wounded area or protruded from the border of the wound were visualized and photographed under an inverted microscope. All the images were collected and analyzed with Image J Tool.
Invasion assay
100,000 cells were seeded in 100 μL of DMEM with 1% of FBS in the upper chamber of the transwell cell culture inserts (pore size, 8 μm; Sarstedt, Numbrecht, Germany). The bottom chamber was added with complete medium (DMEM containing 10% FBS) as a chemoattractant. After 24h, the cells were fixed with 3.7% formaldehyde for 2 minutes and washed two times in PBS, then treated with methanol 100% for 20 minutes, then washed again with PBS and stained with Giemsa. In the upper surface of the membranes the cells were removed with a cotton swab. Cells were observed under a microscope and counted in ten predetermined fields.
Colony formation assay
1,000 cells were seeded in six-well plates. After culturing for 14 days in an incubator, the cells were fixed with methanol and stained with crystal violet. The number of colonies was counted under an inverted microscope.
Cell cycle analysis
MCF7-ShTC1, MCF7-ShSYNGR2, MCF7-TC1-Ctrl and MCF7-SYNGR2-Ctrl cells were harvested, washed with cold PBS twice and fixed in 70% ethanol at –20 °C for 1h, at least. The cells were washed twice using PBS and resuspended in PBS containing 1 mg/mL RNase A (Sigma Aldrich, St. Louis, MO, USA), 0,1 % v/v of Triton X-100 (Sigma Aldrich, St. Louis, MO, USA) and 25 ug/ml of Propidium Iodide (Sigma Aldrich, St. Louis, MO, USA). The cells were incubated 30 min in the dark at 4°C. The percentage of cells in different phases of the cell cycle was measured by flow cytometry using CyFlow1 Cube 8 Sorter Flow Cytometer (Sysmex Partec, Gorlitz, Germany). Data analysis was performed using FCS express 4 software (De Novo Software™).
Statistical analysis
All the data were statistically analysed by unpaired Student's t-test.
TC1 and SYNGR2 mRNA expression in human tissues
From the gene panel expression analysis on our group of samples only SYNGR2 and TC1 genes showed a statistical significant trend as shown in Fig 1 and in Fig 2. The mRNA levels of both genes increased in both tumor sample types (TMT-HBC BRCA1/2 and TMT-SBC). Interestingly we observed a statistically significant difference between BRCA1/2 carriers as opposed to the non carriers. In fact SYNGR2 and TC1 genes showed a statistical significant increase of expression in NMT-HBC BRCA1/2 and NMT-H BRCA1/2 samples compared to NMT-SBC and NMT-H (p<0,0001 and p=0,005 respectively; Figure 1 and Figure 2).
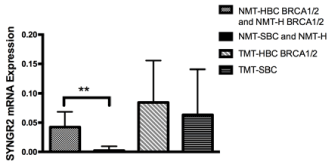
Figure 1. SYNGR2 mRNA expression in patients. The expression in NMT-HBC BRCA1/2 and NMT-H BRCA1/2 was higher than in NMT-SBC and MT-H (p<0,0001). High expression level of the gene is shown in both tumoral patients, BRCA1/2 carriers and not (TMT-HBC BRCA1/2 and TMT-SBC)
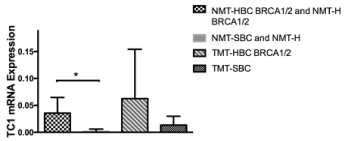
Figure 2. TC1 mRNA expression in patients. The expression in NMT-HBC BRCA1/2 and NMT-H BRCA1/2 was higher than in NMT-SBC and NMT-H (p=0,005)
TC1 and SYNGR2 silencing in MCF7 cells
To evaluate the involvement of SYNGR2 and TC1 genes in breast cancer, we silenced the two genes in MCF7 cells creating stable MCF7-ShSYNGR2 and MCF7-ShTC1 cells. The silencing efficiency of SYNGR2 and TC1 mRNA was evaluated with real time PCR, with a reduction of about 88.4% and 68.8 % respectively (Figure 3). The silencing at the protein level was analyzed with western blot and immunocytochemistry (Figure 4 and Figure 5). The western blot analysis revealed that SYNGR2 protein was 0.39-fold lower in MCF7-ShSYNGR2 cells than in MCF7-SYNGR2-Ctrl cells and TC1 protein was 0.58-fold lower in MCF7-ShTC1 cells than in the negative control cells (Figure 4). Indeed immunocytochemistry analysis confirmed the silencing of the two genes with a protein reduction of 28.3 % for SYNGR2 (p<0.001) (Figure 5a and 5b) and of 42.2 % for TC1 (p=0.0006) (Figure 5c and 5d).
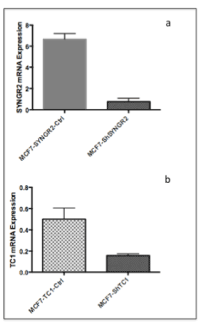
Figure 3. SYNGR2 and TC1 mRNa expression after silencing in MCF7. a) in MCF7-ShSYNGR2 the expression level of SYNGR2 is 88.4% reduced; b) in MCF7-ShTC1 the expression level of TC1 is 68.8 % reduced
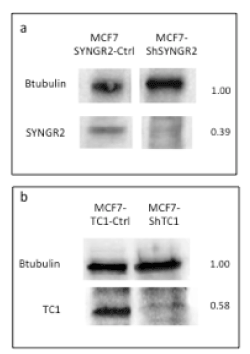
Figure 4. Western Blot Analysis. In a) is shown a reduction of 0.39-fold in MCF7-ShSYNGR2; in b) a reduction of 0.58-fold in MCF7-ShTC1is observed
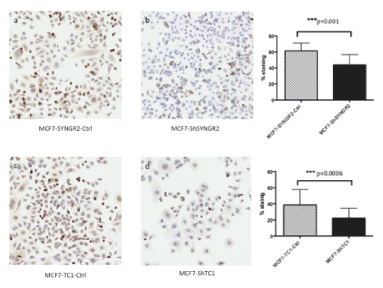
Figure 5. Immunocytochemistry on MCF7-ShTC1 and MCF7-ShSYNGR2 and the negative control cells. In a) and b) is shown the SYNGR2 staining in control cells and in MCF7-ShSYNGR2 respectively. SYNGR2 protein is reduced of 28.3% (p<0.001). In c) and d) is shown immunocytochemistry for TC1 antibody, in MCF7-TC1-Ctrl and in TC1 silencing cell line respectively. TC1 protein is reduced of 42.2% (p=0.0006)
Cellular viability
To evaluate if TC1 and SYNGR2 genes were involved in cellular viability we performed a WST1 assay after 24 hr from transfection. The results revealed that both SYNGR2 and TC1 silencing reduced the viability in MCF7-ShSYNGR2 cells (p=0.035) (Figure 6A) and in MCF7-ShTC1 cells (p=0.0004) (Figure 6B).
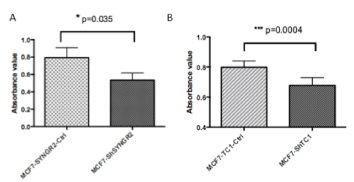
Figure 6. WST1 assay. In a) is shown the viability assay at 24 h for MCF7-ShSYNGR2 and the relative control cells, a reduction of cell viability is observed after SYNGR2 silencing (p=0.035). In b) the viability assay at 24 for MCF7-ShTC1 and MCF7-TC1-Ctrl is shown, TC1 silencing is linked to a cell viability reduction (p=0.0004)
Wound healing assay
To determine if TC1 and SYNGR2 were involved in mediating the migration of breast cancer cells, we evaluated wound healing assay after 24 hours from the scratch. The results showed that the migration of MCF7-ShSYNGR2 and MCF7-ShTC1 cells appeared slower compared to the control even if without a statistical significance (Figure 7a and 7b respectively).
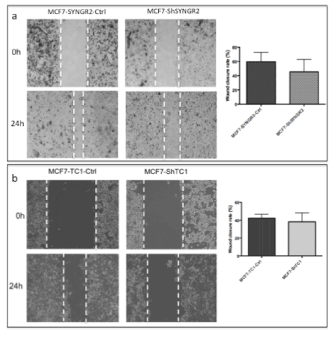
Figure 7. Wound healing assay. In a) is shown the image of the wound after 24 h from the scratch in MCF7-ShSYNGR2 and in MCF7-SYNGR2-Ctrl. A lower migration is characteristic of the MCF7-ShSYNGR2, and in the histogram we can see the percentage of wound closure obtained from analysis with Image J Tool of the different fields; in b) is shown the wound closure after 24 h in MCF7-ShTC1 and in the control cells. A slowdown in the migration after TC1 silencing is detectable
Invasion assay
We observe the transwell chamber after 24 hours of growth, counting the cells of 10 fields for chamber and analyzing them with Image J tool. MCF7-ShSYNGR2 cells invaded slower than the control MCF7-SYNGR2-Ctrl cells without reaching statistical significance (Figure 8a). On the other hand the reduction in invasion of MCF7-ShTC1 compared to control cells was highly significant, as shown in figure 8b (p< 0.0001).
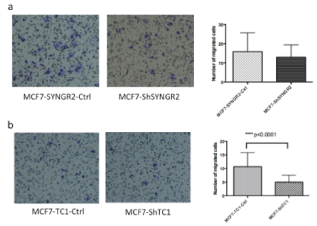
Figure 8. Invasion assay. In the images are shown the membrane of the trans well chamber after 24 h of growth. In a) MCF7-ShSYNGR2 and MCF7-SYNGR2-Ctrl cellular invasion is shown, in the histogram is presented the lower invasion for the MCF7-ShSYNGR2 cells; in b) MCF7-ShTC1 and the control cells are shown, the histogram displays a lower invasion for MCF7-ShTC1 (p<0.0001)
Colony formation assay
As shown in figure 9, SYNGR2 silencing in MCF7 cells caused a reduction in the number of the colonies after 2 weeks compared to the MCF7-SYNGR2-Ctrl (p=0.0193). Silencing of TC1 in MCF7 cells did not cause any statistical difference in colony formation (data not shown).
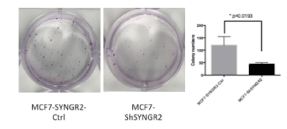
Figure 9. Colony Formation Assay. In the Figure the colonies formated during the assay for MCF7-ShSYNGR2 and MCF7-SYNGR2-Ctrl are shown. A lower presence of colonies in MCF7-ShSYNGR2 is seen rather than in the control cells (p=0.0193)
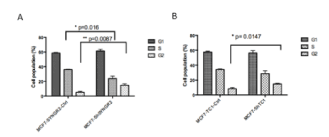
Figure 10. Cell cycle analysis. In picture A) cell cycle of MCF7-ShSYNGR2 and of the respective control cells is shown. The cell percentage of G2/M phase was increased from 5.21 ± 1.27 % in MCF7-SYNGR2-Ctrl cells to 14.8 ± 1.89 % in MCF7-ShSYNGR2 cells (p=0.0087) and the S phase was decreased from 36.07 ± 0.55 % in MCF7-SYNGR2-Ctrl cells to 23.85 ± 3.34 % in MCF7-ShSYNGR2 cells (p=0.016). In B) the cell cycle of MCF7-ShTC1 is compared to MCF7-TC1-Ctrl. The cell percentage of G2/M phase was 8.26 ± 1.61 % in MCF7-TC1-Ctrl cells and 14.8 ± 0.86 % in MCF7-ShTC1 cells (p=0.0147). The S phase was decreased from 34.14 ± 1.1 % in MCF7-TC1-Ctrl cells to 28.65 ± 3.83 % in MCF7-ShTC1 cells
Cell cycle
Flow cytometry was used to detect the effect of SYNGR2 and TC1 silencing on cell cycle progression,. As is shown in Fig 10A, for SYNGR2, the cell percentage of G2/M phase increased of 9.6% going from 5.21 ± 1.27 % in MCF7-SYNGR2-Ctrl cells to 14.8 ± 1.89 % in MCF7-ShSYNGR2 cells and the S phase decreased of 12.2% groing from 36.07 ± 0.55 % in MCF7-SYNGR2-Ctrl cells to 23.85 ± 3.34 % in MCF7-ShSYNGR2 cells. Therefore, SYNGR2 silencing could significantly block the cell cycle progression (p= 0.0087) of MCF7 cells. At the same time, for TC1, the cell percentage of G2/M phase had an increase of 6.5 %from 8.26 ± 1.61 % in MCF7-TC1-Ctrl cells to 14.8 ± 0.86 % in MCF7-ShTC1 cells. By contrast, the cell percentage of S phase decreased of 5.5% from 34.14 ± 1.1 % in MCF7-TC1-Ctrl cells to 28.65 ± 3.83 % in MCF7-ShTC1 cells. This indicates that also the silencing of TC1 could strongly block (p = 0.014) the cell cycle progression of MCF7 cells.
One of the main characteristic of breast cancer is the classification in multiple histopathological subtypes, with different prognosis and response to therapy despite an apparently homogeneous morphology [7,8]. As regards HBC, the identification of the germline mutations in BRCA1 and BRCA2 genes, modified the clinical approach to these disorder. But this is not enough to explain the inter-individual variability in cancer risk. As early as 2001, Hedenfalk et al. demonstrated that different groups of genes were expressed by HBCs BRCA1/2 mutation carriers and SBCs, and even different between BRCA1 mutation carrier tumors and BRCA2 mutation carrier ones [9]. As a matter of fact, modifiers for breast cancer risk can be represented not only by SNPs and mutations, that are commonly studied [10] but also by copy number variations (CNV), miRNA and different gene expression levels [11-13]. Although, in literature, a significant number of microarray profiling studies of breast cancer has been published, only few analyzing HBC exist. This is due to the small group of patients analyzed, in view of the fact that HBC accounts for only a modest part of the breast cancer and also because of the lack of fresh frozen tissues. The vast majority of publications tries to subclassify HBC by molecular profiling [14-15].
We decided to test a 9 genes panel available in our lab proven to discriminate significantly thyroid malignant tumors from benign ones improving considerably cytological diagnostics of fine needle aspirations. The panel is composed mostly of genes known to be associated to cancer including several genes specifically involved in breast cancer such as: KLF4, cKIT and KLF4. Among the nine genes, TC1 and SYNGR2 resulted to have a significant differential expression in the different group of samples that we tested.
TC1 gene, well known in the literature, encodes a small, monomeric protein that functions as a positive regulator of the Wnt/beta-catenin signalling pathway [16]. It was first described overexpressed in papillary thyroid cancer [17]. After that, TC1 has been studied in many human cancers, but with some contrasting results: a large number of studies detected its overexpression in a wide range of tumors [18-21], suggesting that TC1 is involved in aggressive behaviour and poor differentiation for cancer cells. Anyhow, TC1 is downregulated in colon cancer [19] and weakly expressed in hepatocellular carcinoma [22]. About breast tissues, Ray et al. found that TC1 is located on 8p11-p12, that is a breast cancer sensitive genomic region, and that is upregulated in breast cancer cell lines and tissues [23].
The SYNGR2 gene encodes an integral membrane protein containing four transmembrane regions and a C-terminal cytoplasmic tail [24]. It is a member of the synaptogyrin family. The function of this protein is unclear so far, even if it could play a role in regulating membrane traffic in non-neuronal cells [25].
Unlike TC1, there are very few information about SYNGR2 gene and its role, expecially nothing is known about its tumoral implication; it may play a role in regulated exocytosis. In neuronal cells, modulates the localization of synaptophysin/SYP into synaptic-like microvesicles and may therefore play a role in the formation and/or the maturation of this vesicles [24,25].
In our study, we found TC1 and SYNGR2 genes overexpressed in breast cancer tissues (TMT-HBC BRCA1/2 and TMT-SBC) but, intriguingly, also in normal mammary tissues of patients with BRCA1/2 HBC (NMT-HBC BRCA1/2) and in healthy BRCA1/2 mutation carriers (NMT-H BRCA1/2) compared to the normal mammary tissues of cancer patients with SBC and healthy patients without the BRCA1/2 mutations (NMT-SBC and NMT-H).
Since we found TC1 and SYNGR2 upregulated in both SBC and HBC we decided to confirm their involvement in maintaining the tumoral behaviour, by silencing their expression in MCF7 breast cancer cell line. Therefore, we created two permanent TC1- and SYNGR2-silenced cell lines.
TC1 suppression in MCF7 cells showed a reduction in cell viability and invasion as it is reported also by Su et al. in 2013 [21] in A549 lung adenocarcinoma cell line and a decrease in cell migration, even if not statistically significant. The suppression of SYNGR2 gene, never performed before, led to a decrease in cell viability, and in cellular migration and in colony formation. We observed also a reduction of cellular invasion, even if it did not reach statistical significance.
The cell cycle analysis revealed for both MCF7 cell lines silenced for SYNGR2 and TC1 an arrest at G2/M transition with a consequent block of cell proliferation.
The results obtaining from functional studies confirm what is reported by numerous papers about TC1 gene and its involvement in breast cancer aggressiveness. SYNGR2, which is currently a gene without a clear function, could be considered a new potential oncogene. As a matter of fact, this is the first time that SYNGR2 is described involved in a malignant process.
The key finding of our work is the fact that two potential oncogenes are found significantly over expressed not only in the normal tissues of HBC patients but also in the normal tissues of healthy BRCA1/2 mutation carriers, while no expression is found in the normal tissues of SBC. All this indicates that these two genes could contribute to reinforce a molecular profile already prone to the develop of the cancer and could be potential new therapeutic targets to exploit. Moreover, the study of the expression of the TC1 and SYNGR2 gene in a much wider casistic could lead to a better management of the individuals carrying mutations in the BRCA1/2 genes. Normal breast tissue of BRCA1/2 carriers could be another source for the discovery of other genetic determinants that promote the onset of HBC. In conclusion, we propose to look for hereditary cancer risk modifiers in the gene expression profile of BRCA1/2 carriers normal breast tissue, thus integrating the mutational screening approach of germline DNA.
Chiara Maria Mazzanti, Sara Franceschi, Cristian Scatena, Michele Menicagli, Paolo Aretini, Prospero Civita, Claudia Scopelliti, Sara Tomei, Mariella Tancredi, Antonio Giuseppe Naccarato, Maria Adelaide Caligo, Francesca Lessi
The authors declare that they have no competing interests.
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, et al. (2015) Global cancer statistics, 2012. CA Cancer J Clin 65: 87-108. [Crossref]
- Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, et al. (2003) Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet 72: 1117-1130. [Crossref]
- Friebel TM, Domchek SM, Rebbeck TR (2014) Modifiers of cancer risk in BRCA1 and BRCA2 mutation carriers: systematic review and meta-analysis. J Natl Cancer Inst 106: dju091. [Crossref]
- Chenevix-Trench G, Milne RL, Antoniou AC, Couch FJ, Easton DF, et al. (2007) An international initiative to identify genetic modifiers of cancer risk in BRCA1 and BRCA2 mutation carriers: The consortium of investigators of modifiers of BRCA1 and BRCA2 (CIMBA). Breast Cancer Res 9: 104.
- Tomei S, Marchetti I, Zavaglia K, Lessi F, Apollo A, et al. (2012) A molecular computational model improves the preoperative diagnosis of thyroid nodules. BMC Cancer 12: 396.
- Mazzanti C, Zeiger MA, Costouros NG, Umbricht C, Westra WH, et al. (2004) Using gene expression profiling to differentiate benign versus malignant thyroid tumors. Cancer Res 64: 2898-2903. [Crossref]
- Rakha EA, Reis-Filho JS, Ellis IO (2008) Basal-like breast cancer: a critical review. J Clin Oncol 26: 2568-2581. [Crossref]
- Alizadeh AA, Ross DT, Perou CM, van de Rijn M (2001) Towards a novel classification of human malignancies based on gene expression patterns. J Pathol 195: 41-52. [Crossref]
- Hedenfalk I, Duggan D, Chen Y, Radmacher M, Bittner M, et al. (2001) Gene-expression profiles in hereditary breast cancer. N Engl J Med 344: 539-548. [Crossref]
- Walker LC, Marquart L, Pearson JF, Wiggins GA, O'Mara TA, et al. (2017) Evaluation of copy-number variants as modifiers of breast and ovarian cancer risk for BRCA1 pathogenic variant carriers. Eur J Hum Genet 25: 432-438.
- Pinto R, De Summa S, Pilato B, Tommasi S (2014) DNA methylation and miRNAs regulation in hereditary breast cancer: epigenetic changes, players in transcriptional and post- transcriptional regulation in hereditary breast cancer. Curr Mol Med 14: 45-57.
- Forma E, Krzeslak A, Bernaciak M, Romanowicz-Makowska H, Brys M (2012) Expression of TopBP1 in hereditary breast cancer. Mol Biol Rep 39: 7795-7804. [Crossref]
- Tanic M, Yanowski K, Gómez-López G, Rodriguez-Pinilla MS, Marquez-Rodas I, et al. (2015) MicroRNA expression signatures for the prediction of BRCA1/2 mutation-associated hereditary breast cancer in paraffin-embedded formalin-fixed breast tumors. Int J Cancer 136: 593-602. [Crossref]
- Larsen MJ, Kruse TA, Tan Q, Laenkholm AV, Bak M, et al. (2013) Classifications within molecular subtypes enables identification of BRCA1/BRCA2 mutation carriers by RNA tumor profiling. PLoS One 8: e64268.
- Lisowska KM, Dudaladava V, Jarzab M, Huzarski T, Chmielik E, et al. (2011) BRCA1-related gene signature in breast cancer: the role of ER status and molecular type. Front Biosci (Elite Ed) 3: 125-136. [Crossref]
- Jung Y, Bang S, Choi K, Kim E, Kim Y, et al. (2006) TC1 (C8orf4) enhances the Wnt/beta-catenin pathway by relieving antagonistic activity of Chibby. Cancer Res 66: 723-728.
- Chua EL, Young L, Wu WM, Turtle JR, Dong Q (2000) Cloning of TC-1 (C8orf4), a novel gene found to be overexpressed in thyroid cancer. Genomics 69: 342-347. [Crossref]
- Kim B, Koo H, Yang S, Bang S, Jung Y, et al. (2006) TC1(C8orf4) correlates with Wnt/beta-catenin target genes and aggressive biological behavior in gastric cancer. Clin Cancer Res 12: 3541-3548.
- Friedman JB, Brunschwig EB, Platzer P, Wilson K, Markowitz SD (2004) C8orf4 is a transforming growth factor B induced transcript downregulated in metastatic colon cancer. Int J Cancer 111: 72-75. [Crossref]
- Yang ZQ, Moffa AB, Haddad R, Streicher KL, Ethier SP (2007) Transforming properties of TC-1 in human breast cancer: interaction with FGFR2 and beta-catenin signaling pathways. Int J Cancer 121: 1265-1273. [Crossref]
- Su K, Huang L, Li W, Yan X, Li X, et al. (2013) TC-1 (c8orf4) enhances aggressive biologic behavior in lung cancer through the Wnt/β-catenin pathway. J Surg Res 185: 255-263. [Crossref]
- Zhu P, Wang Y, Du Y, He L, Huang G, et al. (2015) C8orf4 negatively regulates self-renewal of liver cancer stem cells via suppression of NOTCH2 signalling. Nat Commun 6: 7122. [Crossref]
- Ray ME, Yang ZQ, Albertson D, Kleer CG, Washburn JG, et al. (2004) Genomic and expression analysis of the 8p11-12 amplicon in human breast cancer cell lines. Cancer Res 64: 40-47. [Crossref]
- Belfort GM, Kandror KV (2003) Cellugyrin and synaptogyrin facilitate targeting of synaptophysin to a ubiquitous synaptic vesicle-sized compartment in PC12 cells. J Biol Chem 278: 47971-47978. [Crossref]
- Belfort GM, Bakirtzi K, Kandror KV (2005) Cellugyrin induces biogenesis of synaptic-like microvesicles in PC12 cells. J Biol Chem 280: 7262-7272. [Crossref]










