Abstract
We established and operated a laboratory associated with the Ebola Treatment Center of Macenta between November 2014 and December 2015. The laboratory was designed to ensure a high level of biosafety and assurance quality and provide physicians with diagnoses for Ebola virus disease (EVD), as well as Lassa fever, malaria, and typhoid fever. We included equipment that permitted basic biochemistry analyses to improve the supportive care of patients. Once the outbreak of EVD was over in forested Guinea, the laboratory remained involved in the epidemiological surveillance of EVD among community deaths of the entire area and participated in clinical research projects. The lessons learned from this experience were that giving priority to biosafety and assurance quality was critical and providing biochemistry analyses, in addition to etiological diagnoses, is becoming essential in such a field lab. Among the challenges we had to face, the most problematical issues were insuring a continuous and reliable supply of reagents and consumables and having a sufficient pool of skilled staff to allow team rotations.
Keywords:
Ebola; Guinea
Introduction
In the summer of 2014, several Ebola treatment centers (ETCs) were urgently established to respond to the increasing number of Ebola virus disease (EVD) cases in Guinea [1]. One of these centers was set up in Macenta, a town of 45,000 inhabitants, located in South-East Guinea, close to the borders with Sierra Leone and Liberia. The ETC was funded by the French Ministry of Foreign Affairs, set up by Médecins Sans Frontières, and operated by the French Red Cross, with human resources support from the Etablissement de Préparation et de Réponse aux Urgences Sanitaires (EPRUS, French Ministry of Health). The Institut Pasteur set up and ran a laboratory, close to the ETC, to provide diagnostic capacity near the treatment center. The Pasteur Laboratory also handled the samples from the ETC at Nzerekore, which was operated by the Alliance for International Medical Action (ALIMA), until the opening of a laboratory associated with this ETC. In addition, we also received samples from different hospitals in the area. The laboratory opened on November 29th, 2014 and definitively closed and was dismantled in mid-December, 2015. During this period, 564 samples from 422 patients were processed in the laboratory. Most of the EVD patients were diagnosed during the five weeks following the opening of the laboratory, and a small cluster of patients was identified in the first weeks of 2015 (Figure 1). The ETC of Macenta was empty thereafter and the activity of the laboratory focused on the diagnosis of Ebola virus (EBOV) infection among communitarian deaths and participation in the POSTEBOGUI project (IRD-INSERM), with the aim of studying the persistence of EBOV within various biological fluids of EVD survivors [2]. The role of the laboratory was crucial during this period, as it was the last active laboratory in forested Guinea. It was operated by teams composed of three people, including two technicians and one biologist, the latter being the head of the team. Teams worked seven days a week, for three to four weeks, and were then replaced by another team with a few overlapping days. The equipment of the laboratory was funded by the Agence Française de Développement, and the design and set-up of the laboratory was performed by the authors.
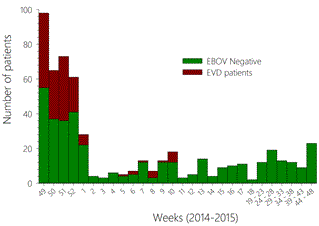
Figure 1. The number of patients for whom samples were analyzed is presented according to the 2014 - 2015 weeks. The patients excluded for EBOV infection are presented in red whereas confirmed EVD patients are in green.
Laboratory Concept
We were asked to establish a laboratory that would operate for at least one year. Thus, we choose to set-up a laboratory at a fixed site, rather than a mobile one, to ensure long-term functionality of the equipment. Moreover, the laboratory was designed with an emphasis on quality and biosafety. The laboratory was established in a three-room building to ensure an optimal RT-PCR pathway (Figure 2). Room #1 was reserved for the handling of infectious materials in a four-gloves biological safety cabinet class III (BSCC3) (Eurobioconcept, Bonneuil sur Marne, France) and storage of infectious samples. Room #2 was used for RNA extraction, preparation of RT-PCR mixtures, and reagent storage. Finally, room #3 was simultaneously used for amplifications and as the office.
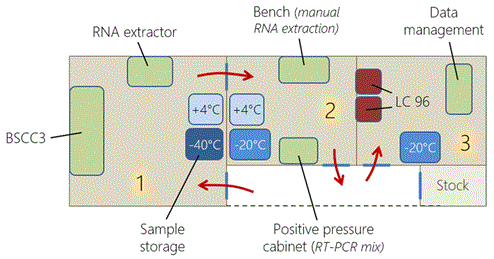
Figure 2. Three rooms were used in the laboratory and a fourth room was used to store consumables (stock). There was one BSCC3, on RNA extractor, two refrigerators (+4°C), three freezers (-20°C and -40°C), one positive pressure cabinet, two LightCycler 96 thermocyclers. The red arrows indicate the way of moving in the successive rooms.
Provision of quality assurance compatible with medical biology needs
Medical biology and diagnostic activities require a robust quality management system (QMS) to ensure the delivery of reliable results. This is particularly true for EVD, a disease for which false negative or positive results cannot be tolerated. Indeed, the consequences of either mistake are tragic. False negative results would lead to EBOV-infected patients leaving the ETC and transmitting the disease to their relatives or health-care workers, whereas false positive results would result in infection by EBOV by the patient within the ETC. These risks are not theoretical, as both situations occurred during the West African outbreak. The QMS was applied to the various steps of the diagnosis process, including human resources, and will be described in the following paragraphs.
Provision of optimal biosafety despite field conditions
Handling EBOV requires a BSL4 laboratory or at least a level-3 safety cabinet and stringent rules and processes for optimal safety during sample handling and waste disposal. We choose to install a BSCC3 equipped with a double door entrance and two exits with double-doors for leak tight transfer (DDLT), one for waste disposal using heat-sealing bags and the other for the transfer of materials or samples. This glove-box was operated under 200 Pa negative pressures and sterilized once a day using vaporized peracetic acid. Materials and disposables used within the glove box and solid waste were decontaminated using 0.5% sodium hypochlorite, followed by peracetic acid, before exiting. We choose to use 0.5% sodium hypochlorite as a disinfectant because of its demonstrated efficiency against EBOV [3]. Waste-containing bags were immediately incinerated. Personal protective equipment (PPE) was used by the staff to work in room 1, despite the infectious materials being handled exclusively within the glove box (Figure 3). This equipment was justified by a critical step which was the transfer of infectious samples contained on a single cover-slip from the glove box to the -40°C freezer. Long-term storage of single cover-slips of infectious samples was not allowed in the freezer and EBOV-positive samples were packed according to UN 6.2 instructions as soon as the results of the diagnosis were known. This process avoids further handling of infectious samples for shipment. To ensure optimal and long-term compliance with the biosafety-related instructions, all processes were exhaustively listed in written procedures.

Figure 3. The PPE worn by staff to work in room #1 is presented. These equipment were discarded before entering into room #2 and replaced by a disposable overall and a single pair of gloves.
Provision of differential diagnoses and biochemistry analyses to improve patient care
The evolution rate of EBOV after multiple human-to-human passages was unknown at the time we designed the laboratory. We thus chose to use the Realstar Filovirus screen kit 1.0 (Altona Diagnostics, Hamburg, Germany) to ensure the detection of the virus even if it evolved during circulation, as this assay was designed for all EBOV species [4]. This assay is less sensitive than the Zaire EBOV-specific RT-PCR, but this was not a concern as all the diagnoses were performed on acutely-ill patients, with a presumably high viral load. The sensitivity of the assay, demonstrated in the field, was sufficient (500-3,000 RNA copies/mL) [4]. We also performed rapid diagnostic tests for malaria and typhoid fever (ONSITE PF/pan Malaria Ag Rapid Test and ONSITE Typhoid IgG/IgM Combo Rapid Test/K7, both from CTK Biotech, San Diego, CA), because of the high prevalence of these diseases in the area. A home-made real-time RT-PCR assay targeting the glycoprotein precursor gene of Lassa virus was also available in the laboratory, as Lassa fever is endemic in the area and induces similar symptoms as EVD. This latter diagnosis was performed retrospectively, but regularly, on samples from patients who were found to be negative for EVD, malaria, and typhoid fever, but presented a clinical picture consistent with Lassa fever.
We also helped the physicians in the supportive care provided within the ETC to EVD patients, by performing basic biochemical analyses of patient samples using a Piccolo Xpress (Abaxis, Union City, CA) and two i-stats analyzers (Abbott). We chose this equipment because of its small size and the absence of fluid circulation inside the equipment, limiting internal contamination of the instrument. The panels used were Amlyte 13 for Piccolo and Chem8+ for i-Stat. The combination of these two panels provided useful basic data concerning electrolytes, hepatic, and renal function, as well as inflammatory markers, including: Ca2+, Cl-, Na+, K+, AST, ALT, amylase, bilirubin, urea, creatinine, creatine-phosphokinase, C-reactive protein, glucose, albumin, and hemoglobin.
Preanalytical Steps
During the period of the epidemic, the laboratory received samples from the ETC of Macenta, the Guinean Red Cross handling the community deaths, and regional hospitals. We were also in charge of the samples coming from the ETC of Nzerekore (operated by the Alliance for International Medical Action) during the first two months of operations. When the outbreak was over in forested Guinea, the other laboratories closed and the Pasteur laboratory was the last to operate in the area. Consequently, the laboratory received all samples coming from hospitals and community deaths.
Blood samples were harvested using EDTA, for etiological diagnoses, and heparin for biochemistry analyses. Saliva swabs were also handled for EVD diagnoses of young children or dead patients. We engaged to provide an etiological diagnosis result within five hours of reception, which was the shortest time possible considering sample processing in the BSCC3 and the subsequent RT-PCR. The samples arrived in the laboratory together with an administrative and clinical data sheet (Figure 4). The administrative and clinical details of the patient were included, with an emphasis on the date of symptom onset, which was crucial to interpret the EBOV RT-PCR results. A unique identification number was then attributed to each sample and used to complete the sample register (also containing the patient’s name and age, date of sampling and origin, initial ETC identification number, type of sample, requested analyses, and operator signature) and label the various tubes to be used. The match between samples and the completed forms was verified using a double reading procedure to prevent mistakes. This step was crucial, as common given names are very frequent in the area. We also developed a specific Microsoft Access® database that was used to enter administrative, clinical, diagnosis, virological, and biochemistry information and results. It allowed us to link the different samples from the same patient and to monitor the evolution of the disease and the virological parameters. This database is also particularly helpful for clinical research studies on our patients as it allows access to clean and reliable data. Access to the database was restricted to those authorized to validate the biological results.
Figure 4. Fac simile of the form filled by physicians and accompanying patient’s samples. The verso that contained fields filled by the laboratory staff (results, time of experiment…) is not presented.
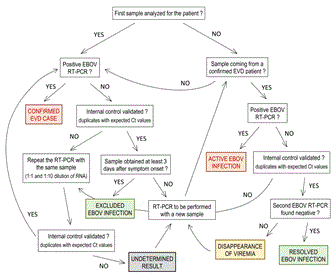
Figure 5. Consort diagram for EBOV diagnosis interpretation and validation. The different possible results are presented. The conclusions are colored in red in case of EBOV infection, in green when a negative EBOV RT-PCR was obtained in a sample coming from an EVD confirmed case, and in grey when the result was undetermined.
Analytical Steps
We performed two series of analyses per day, seven days a week. It is only possible to open the double door entrance of the BSCC3 twice before the need for sterilization. Thus, we performed the second series in the evening. In addition to EBOV RT-PCR, we also performed rapid diagnostic tests (RDT) for malaria and typhoid fever for the initial diagnosis of patients with symptoms consistent with Ebola virus disease for whom a blood sample was available. The malaria RDT could distinguish Plasmodium falciparum infection from other Plasmodium species endemic in the area. We first performed the RDT using 5 and 50 µl of whole blood for malaria and typhoid fever, respectively. Then, heparinized-whole blood, if available, was used for biochemical analyses. Blood samples were centrifuged and the plasma was aliquoted. Plasma from EDTA samples was used for RNA extraction for viral genome detection and the remaining volume stored at -40°C for further experiments and clinical research. We treated the samples of patients who had already been tested at least once in the same manner, except the rapid tests for malaria and typhoid were not performed. There were two possible scenarios: if a patient was found to be negative for EBOV infection using a sample obtained less than three days after the onset of symptoms, a new one was processed at least three days after disease onset to exclude or diagnose EBOV infection. For patients with confirmed EVD, further analyses were performed to monitor the evolution of the viral load and biochemical parameters. When EBOV RNA became undetectable in a sample of an EVD patient, another sample was obtained 48 h later to confirm the disappearance of viremia and allow the patient to be discharged from the ETC. For saliva samples, the swab was soaked and shaken in 300 µl of water. One hundred microliters of this solution was then used for extraction and the remaining volume stored at -40°C for further analyses.
RNA extraction and RT-PCR for EBOV
One hundred microliters sample (EDTA plasma or water from a swab) was inactivated in 400 µl AVL buffer (Qiagen, Courtaboeuf, France) supplemented with 4 µl RNA carrier and the internal control provided in the Realstar Filovirus screen kit 1.0. The internal control allowed validation of the extraction and amplification steps, which was crucial as we frequently observed the inhibition of RT-PCR. Our first aim was to use automated extraction. We chose an EZ1 Advanced XL automated extractor (Qiagen, Hilden, Germany) because it was designed for diagnostic purposes and only needs a single consumable reference. Moreover, 14 samples could be extracted simultaneously. The extraction protocol did not comply with validated EBOV inactivation procedures. We compared the reference method (manual extraction using the Qiamp Viral RNA kit from Qiagen) to the alternative method, including inactivation in AVL buffer followed by extraction using the EZ1 DSP virus kit (Qiagen), in our laboratory in France and found it to be sufficient. However, the extraction efficiency varied somewhat among samples for both methods in the field. We finally chose to extract the RNA using the Qiamp Viral RNA kit to guaranty the best detection sensitivity. A recent study demonstrated that the addition of absolute ethanol is required for complete inactivation of EBOV [5]. We thus updated our procedures with the addition of ethanol 10 minutes after the lysis of plasma in AVL buffer in the BSCC3, impeding the use of the EZ1 RNA extractor. The tubes were soaked for 10 minutes in 0.5% sodium hypochlorite before being removed from the BSCC3. The further steps of the procedure were performed according to the manufacturer’s recommendations in room #2. RT-PCR was then performed using the Realstar Filovirus Screen kit 1.0 (Altona diagnostics). Briefly, the RT-PCR reaction mixture was prepared in positive-pressure cabinet (BioHEPA 321, Captair, Val de Reuil, France) in room #2, and the RNA added to the reaction mixture in the same room, but outside the cabinet. EBOV positive and negative controls, as well as controls of extraction/inhibition, were systematically included in the series. Finally, amplification was performed in room #3 using LightCycler 96 instruments (Roche).
Postanalytical Steps
The results of the analyses, diagnoses and biochemistry, were exclusively processed by the person authorized to validate biological results. This authorization was delivered by the head of the laboratory (S. Baize) based on their training and diploma, experience in medical biology and diagnosis, and specific knowledge of EBOV infection and EVD. Access to the patient database was given after signature of a letter of engagement to comply with the procedures of the laboratory, respect the confidentiality of the patient data, and follow the flowchart used to interpret the diagnostic results. A period of double validation by the head of the laboratory was sometimes established to confirm their skills. After validation, the authorized individuals immediately handed the results over to the medical staff and the consequences of the laboratory results for the patient were systematically discussed between the medical staff and the laboratory.
Validation of the etiological diagnosis
A specific flowchart was used to interpret and validate the results obtained using the Realstar Filovirus Screen kit 1.0 (Figure 5). The use of such a flowchart allowed us to ensure consistent interpretation of the results, despite the frequent changes of laboratory staff. The conclusions ‘confirmed EVD case’ and ‘active EBOV infection’ resulted in admission, and the patient was kept in the ‘confirmed case’ area of the ETC. The conclusion ‘undetermined result’ led to keeping the patient where they already were, either in the ‘probable cases’ area or the ‘confirmed case’ area. The conclusion ‘disappearance of viremia’ also resulted in keeping the patient in the ‘confirmed case’ area. Finally, the conclusion ‘excluded EBOV infection’ or ‘resolved EBOV infection’ led to the discharge of the patient from the ETC. Inhibition of RT-PCR was not rare. When this occurred, the RT-PCR was repeated with the same RNA sample, but using both 1:1 and 1:10 dilutions, as described in the flowchart. If the problem persisted, a new analysis was performed on new sample.
We did not detect any cases of LASV infection, by RT-PCR for the GPC gene of LASV, among the patients admitted to the ETC who were negative for EBOV infection. This is not surprising, as we tested only a small number of patients and no outbreak of Lassa fever occurred in the area during the period. Active typhoid fever is confirmed in patients if specific IgMs are detected by the RDT. We never confirmed typhoid fever in the patients, although we observed a faint IgM band for a few. The RDT for malaria allowed us to discriminate between P. falciparum and other African species. We found a high rate of P. falciparum positive samples among EBOV-positive (18%) and negative patients (20%), but failed to detect other species without coincident P. falciparum infection. The biochemical results were given to the physicians as crude values without interpretation or conclusion, as our staff did not receive a medical biologist diploma.
The conclusions of the etiological diagnosis, the EBOV RT-PCR Ct values for the positive samples, and the biochemical values were entered in the database and a printed or PDF report was transmitted to the medical staff of the ETC of Macenta personally or to other prescribers by email. When the data was transmitted by email, the results were first given over the telephone to allow discussion of their consequences for the patients. Only the person in charge of biological validation was authorized to give a result to physicians.
Biosafety and Biological Security Procedures
One of the most important elements in the laboratory was confidence in the level of biosafety. This included the use of a BSCC3 that was properly installed on site by a certified technician and was maintained according to the manufacturer’s instructions. The reception of samples in appropriate packages and decontamination by validated procedures were also crucial biosafety measures. Finally, the extensive use of PPE also played a major role in biosafety. Biological security was also critical, as a high number of EBOV-containing samples were stored in the laboratory.
BSCC3 installation and validation of the sterilization process
Upon reception, the BSCC3 was installed and verified for airtightness by a technician certified by the manufacturer. The sterilization method was validated on site using Geobacillus stearothermophilus. The spores, dried on paper, were placed in different areas of the cabinet and their complete inactivation was verified by culturing spores for seven days at 37°C. This procedure allowed us to define the optimal quantity of peracetic acid to use and to validate the sterilization efficiency.
Personal protective equipment
Handling the infectiou2021 Copyright OAT. All rights reservcient to protect the staff. However, we decided to add PPE, including a Tyvek Classic Plus® suit (DuPont, Wilmington, DE), a waterproof surgical gown, overboots, a FFP3 mask, protective glasses, and two pairs of gloves (fixed to the suit with adhesive tape for the internal pair) to work in room #1. Although all the experimentations with infectious samples were made inside the BSCC3, the transfer of cryotubes containing infectious samples to the -40°C freezer and their temporary storage before safety packaging in sealed plastic bags involved the use of PPE able to protect the staff in case of accidental opening of a cryotube. These PPE were discarded before entering room #2, which was the mandatory exit from room #1.
Infectious sample packaging
The samples provided by the ETC of Macenta and those transported by car did not use the same packaging. For the ETC of Macenta, double packaging was sufficient, given its proximity to the laboratory. The tube was enclosed in an absorbent pocket able to absorb more than 100 ml of fluid that was placed in a hermetic plastic bag. All the packaged samples were soaked in 0.5% sodium hypochlorite and brought to the laboratory. Triple packaging was required for samples transported by car, UN 6.2 packaging was then used. Upon reception, this package was opened in room #1 and the second package (hermetic biotainer) was soaked in 0.5% sodium hypochlorite during opening to prevent any contamination if the inner packaging was broken. The tubes were then transferred into the BSCC3 after verifying their correspondence with the clinical data sheets.
Handling of infectious samples
Every sample was considered to be potentially infectious and was handled in the BSCC3. Everything needed for the experiment was prepared following a check-list and transferred into the cabinet through the double door entrance and then to the working area. The inside door of the entrance was closed before opening the tubes, allowing the entrance to remain safe for a second use. The BSCC3 could therefore be used twice before sterilization. Stickers indicating “fully sterilized”, “double door entrance usable once more” or “entrance contaminated” were affixed to the cabinet to prevent any confusion concerning its use. The consumable and biological waste were decontaminated during the experiment using 0.5% sodium hypochlorite and remained in the cabinet during the sterilization process. Waste was then removed from the BSCC3 in sealed plastic bags and immediately incinerated. All liquid waste containers were filled to a third of the total volume with a 1.5% sodium hypochlorite solution, insuring that liquids were decontaminated without falling below the 0.5% limit.
Evolution of the procedures
During the deployment, a study showing that samples were not fully inactivated by the addition of AVL buffer brought this risk to our attention. We modified our procedures to add the ethanol inside the cabinet instead of in room #2, as the addition of ethanol was required for total inactivation.
Biological security
We applied rigorous rules concerning biological security to ensure the safe storage of infectious samples. Access to room #1 and 2 was restricted to the Pasteur Laboratory staff; only room #3 could host other people. The three rooms of the laboratory were systematically closed when the laboratory staff was absent, and the whole ETC site, including the laboratory, was watched by guards 24 hours a day, seven days a week. Moreover, the -40°C freezer, in which the infectious samples were stored, was secured with a robust padlock.
Challenges and Opportunities
Setting up a laboratory reserved for handling risk-group 4 agents that is expected to be functional for at least one year in a limited amount of time and in a remote area is, of course, highly challenging. We had to cope with various concerns at each step.
Some equipment, such as the level-3 safety cabinet takes time to be delivered. Indeed, this equipment is made on demand by the manufacturer to meet the specifications required by a field laboratory, including that it be simple, robust, and easily maintained by non-specialists. In our case, it took six weeks to obtain the safety cabinet. In addition, its exportation outside Europe is subject to obtaining a ‘double-use equipment exportation license’, because of the bioterrorism risk. This license was delivered in our case by the French authorities. Obtaining this document can also take a substantial amount of time, but we obtained it quickly because of the outbreak emergency. Another major challenge is the transport of the material to the site without damage or delay, once all the equipment and consumables are assembled. Such material is specific, sensitive, and subject to specific regulations for transport, and the operators in charge of this task are not always conscious of this fact. For example, several reagents are considered to be dangerous and can only be shipped by air using cargo aircraft, which are not always available. During the operation of the lab, we faced the loss of reagents or equipment or ran out of critical reagents on several occasions, and even a fire of the warehouse where our reagents were stored in Conakry. All these issues seriously impeded the activity of the laboratory, occasionally to the detriment of the patients. During the year that the laboratory was open, the time between ordering reagents and consumables and shipment to our laboratory in Macenta was approximately two to three months, requiring that we largely anticipate our needs. Another challenge of operating such a laboratory in the field was the requirement of a stable and continuous supply of electricity. In addition to the use of generators, each critical piece of equipment (BSCC3 and its sterilizer, thermocyclers, computers…) requires an uninterruptible power supply to avoid power cuts and voltage variations. Finally, the duplication of critical instruments is essential to manage the failures that will inevitably occur.
The long-term management of human resources was also particularly challenging. The teams were present on site for three weeks before being replaced by a new one. It rapidly became difficult to find new qualified staff. Fortunately, we were able to recruit a Guinean technician who was permanently present, limiting the number of temporary personnel.
The design of the laboratory allowed us to maintain its activity during the entire year of operation and to participate in clinical research projects led in the Macenta area. Two different diagnostic assays were evaluated in the laboratory and importantly, a substantial portion of the samples collected in the cohort of survivors followed by the POSTEBOGUI project has been analyzed in the lab, confirming the persistence of EBOV RNA in semen for up to 9 months after disease onset [2]. Finally, the establishment of a laboratory at a fixed site in a remote area can also help to implement the health-care infrastructures of countries and perform post-epidemic surveillance and follow-up.
Recommendations for Future Deployment
The lessons learned from our experience indicate that establishing a laboratory in a limited amount of time and in a remote area is possible without giving up biosecurity and quality assurance. We did not encounter a single issue regarding biosafety during the entire period that the laboratory was open, and to our knowledge, no diagnostic results of EBOV infection have been challenged. The access to supplementary analyses in addition to diagnosis, including biochemical analyses, is becoming necessary, and the possibility to perform hematological analyses is advisable. Indeed, the input of the laboratory in this matter is of prime importance as improved survival is correlated with improved supportive care. Concerning differential diagnoses, the diagnosis of malaria is particularly crucial, as a considerable portion of patients were infected, independent of their EBOV status. The administration of rapid malaria therapy is therefore crucial and can decrease overall mortality. Typhoid serology was, ultimately, not informative. Very few patients were IgM positive and this was not a clear sign of present infection. No antigenic rapid test was commercially available during this period, and this diagnosis could be revisited in the future if new RDT based on antigen detection becomes available. It is important to still test for LASV given the endemicity of Lassa fever in West Africa and the similarity of its clinical symptoms with those of EBOV, even though no case of LASV infection was detected in our cohort. Indeed, a high number of Lassa fever cases were consistently detected in other ETCs, notably in Sierra Leone. Finally, some aspects must be improved for future deployment, such as the late establishment of the lab, only two months before the end of the outbreak in forested Guinea, and the reliable supply of reagents and consumables, that was the biggest problem.
References
- WHO Ebola Response Team (2014) Ebola virus disease in West Africa--the first 9 months of the epidemic and forward projections. N Engl J Med 371: 1481-1495. [crossref]
- Sow MS, Etard J-F, Baize S, Magassouba NF, Faye O, et al. (2016) New Evidence of Long-lasting Persistence of Ebola Virus Genetic Material in Semen of Survivors. J Infect Dis. [crossref]
- Cook BW, Cutts TA, Nikiforuk AM, Poliquin PG, Court DA, et al. (2015) Evaluating Environmental Persistence and Disinfection of the Ebola Virus Makona Variant. Viruses 7: 1975-86.
- Rieger T, Kerber R, El Halas H, Pallasch E, Duraffour S, et al. (2016) Evaluation of RealStar Reverse Transcription–Polymerase Chain Reaction Kits for Filovirus Detection in the Laboratory and Field. J Infec Dis 214: S243-S249.
- Smither SJ, Weller SA, Phelps A, Eastaugh L, Ngugi S, et al. (2015) Buffer AVL Alone Does Not Inactivate Ebola Virus in a Representative Clinical Sample Type. J Clin Microbiol 53: 3148-3154. [crossref]





