Background: Unequivocal diagnosis of FSGS can only be made with a renal biopsy, which is an invasive, risk-associated medical procedure. The discovery of non-invasive molecular biomarkers for the diagnosis of FSGS remains an important scientific goal. This study examines the urinary proteome of FSGS patients and reference groups, in order to identify urinary protein expression alterations indicative of FSGS.
Methods: Urine samples were collected from subjects representing FSGS, IgA nephropathy (IgAN), clear cell renal cell carcinoma (ccRCC), chromophobe renal cell carcinoma (chRCC), and healthy control group, respectively. The samples were subjected to SWATH-MS proteomics analysis. ELISA was utilized to validate the expression level of Retinol-binding protein 4 (uRBP4) in FSGS and reference samples (IgAN, ccRCC, chRCC, prostate cancer and healthy subjects).
Results: The MS study identified 194 (FSGS), 179 (IgAN), 271 (ccRCC), 255 (chRCC), and 275 (healthy controls) urinary proteins. The comparative proteomic analysis revealed that urinary Retinol-binding protein 4 (uRBP4) clearly discriminates FSGS from the rest of the groups. Increased levels of uRBP4 in FSGS urine specimens were also evidenced by ELISA. Significantly elevated levels of uRBP4 were also observed for IgAN, ccRCC and chRCC versus healthy individuals.
Conclusions: Determining FSGS diagnosis based on uRBP4 expression alone is not possible. Specific uRBP4 concentration cut-off can be applied to accurately distinguish individuals with renal disorder (in general) from healthy subjects. Possibly, urinary RBP4 could serve as a screening biomarker identifying people at risk of renal disorders, who should undergo more detailed diagnostics.
FSGS, renal diseases, urinary RBP4
Focal segmental glomerulosclerosis is the predominant subtype of primary glomerular diseases (primary glomerulopathies, PGs) in adults and the commonest cause of chronic renal failure (chronic kidney disease, CKD) [1]. The worldwide incidence of this rare condition is 0.8 cases per 100,000 subjects per year [2]. The precise diagnosis of FSGS is based entirely on kidney biopsy that is an aggressive procedure associated with risk of complications [1]. Presently, there are no molecular diagnostic strategies available for FSGS that could serve as a non-invasive alternative to renal biopsy. Undoubtedly, urinary proteome comprises a valuable source of biomarkers of kidney diseases, as much of the proteins come directly from the kidneys and reflect their actual physiological status. Urinary proteins are relatively stable comparing to the proteins of sera, and are more desired as biomarkers due to a non-invasive way of urine collection [3]. Interestingly, urinary proteins proposed hitherto as possible candidates of chronic kidney diseases seemed to outperform the ‘biomarkers’ found in the blood [4].
There is a number of studies comparing the pattern of urinary proteome of FSGS patients with the proteomic profiles of healthy controls and/or patients with other renal-diseases [5-11]. However, the past research have never confronted simultaneously the pattern of urinary proteome in FSGS patients with the proteomic profile of the normal status, IgA nephropathy (the second most common glomerulopathy worldwide) and the commonest subtypes of renal cancers i.e. ccRCC and chRCC. Such strategy i.e. to mine and compare the urinary proteomes of distinct renal disease states (glomerulopathies, kidney cancers, normal status) may enhance the chances of finding quantitative/qualitative proteomic mark truly specific for particular renal disorder. Within the framework of this study we aimed to characterize the urinary proteome of FSGS subjects, and compare it to the protein expression profiles of the following reference groups: healthy controls, IgAN subjects, ccRCC, chRCC, and prostate cancer subjects in order to extract proteins specifically indicative of the FSGS.
Ethics statement
This study was approved by the Bioethics Committee of Regional Specialized Hospital in Wrocław (approval ID: KB/nr 9/rok 2016). All samples were collected at Regional Specialized Hospital after obtaining written informed consent from study participants.
Description of participants
Participants in this study included healthy volunteers (age range; 23 – 60, n = 30, 16 men and 14 women), patients with biopsy-proven primary FSGS (age range 24-74, n = 20, 16 men and 4 women), and primary IgAN (age range 21-66, n = 19, 11 men and 8 women), patients with histopathologically confirmed primary ccRCC (age range 48-89, n = 31, 18 men and 13 women), primary chRCC (age range 19-86, n = 21, 12 men and 9 women), and prostate cancer (age range 57-79, n = 7 men).
Urine sample collection and handling
All participants provided first-morning urine (20 ml - 50 ml) in a sterile urine containers. Urine samples were obtained in the morning before scheduled renal biopsy (IgAN, FSGS patients) or surgery (renal cancer and prostate cancer patients). The specimens were processed at room temperature within 1-3 hours post-collection. All samples were tested for the following parameters: leukocyte esterase, nitrite, urobilinogen, protein, pH, blood, specific gravity, ketone, bilirubin, glucose with a Multistix 10 SG Reagent Strips (Siemens Healthcare Diagnostics Inc, NY, USA). Urine specimens of healthy controls and prostate cancer patients had normal urine parameters. The summary of urine dipstick test results is included in Supplementary Table 1. Urine samples were centrifuged at 4300g for 30 minutes at room temperature and the supernatants were aliquoted into 2 ml sterile LoBind tubes, and stored at -80oC until further processing.
Sample preparation for SWATH-MS/MS (quantitative analysis)
Urine supernatants were thawed in a waterbath (37oC) and vigorously vortexed. Equal volumes of urine supernatant from four subjects from the same group were pooled. The pooled samples were concentrated and desalted using 10-kDa molecular weight cutoff membranes (Merck KGaA, Darmstadt, Germany). Concentrated proteins were washed twice and eluted with equal volumes of molecular biology grade water (4000g/room temperature/20 minutes). The protein quantity in each sample was assessed in replicates by Qubit 2.0 fluorometer using Qubit Protein Assay Kit (Thermo Fisher Scientific, MA, USA). Adequate volume of each pooled sample (corresponding to 15µg of proteins) was vacuum dried using speedvac (miVac, Genevac, UK). The protein pellets were suspended in 15 µl 50 mM NH4HCO3 and sonicated in a water bath (15 minutes/no heating). Samples were reduced by 10 mM dithiotreitol (DTT) at 56°C for 30 min, and subsequently alkylated by iodoacetamide in the final concentration of 20 mM (darkness/room temperature/30 minutes). After adding 20 µl of 50 mM NH4HCO3 to each sample, the samples were incubated overnight with MS-grade trypsin (1:50 trypsin to substrate ratio) with constant shaking (750 rpm) at 37oC. Digestion reaction was quenched by 5% formic acid/50% acetonitrile solution. The samples were desalted using a ZipTip C18 (Millipore) according to manufacturer`s instruction. Peptides were eluted into 50% acetonitrile/0,1% formic acid solution and subjected to LC-MS/MS.
Sample preparation for spectral library generation
The spectral librarywas generated by IDA-MS from theunfractionatedpeptide samples (prepared as in section ‘Sample Preparation for SWATH-MS/MS’) and fractionated peptide specimens.
Fractionated peptide samples were prepared from 120 µg and 100 µg protein pellets using Multiple Affinity Removal System (MARS-14, Agilent Technologies), and following the manufacturers` protocol. The low-abundant (LAP) and the high-abundant protein fraction (HAP) were subjected to proteolytic digestion and desalting as described in the section ‘Sample Preparation for SWATH-MS/MS’.
LC-MS/MS
The samples were separated with the Ekspert MicroLC 200 system (Eksigent, CA, USA) using ChromXP C18CL column (3 µm, 120 A, 150 x 0,5 mm). The injection volume was 5µl. The mobile phases consisted of LC-MS grade formic acid (0,1%) in water [A] and acetonitrile [B]. The separation was performed by a 30 minute gradient (0-2 min 10% B; 2-23 min 10-90% B; 23-28 min 90% B, 28,1-30 min 10% B). Data acquisition was performed with a TripleTOF 5600+ mass spectrometer with a DuoSpray Ion Source inpositive ion mode (AB SCIEX, CA, USA). The microLC-MS/MS system was controlled by AB SCIEX Analyst TF 1.6 software.
- LC-MS/MS in IDA (information-dependent acquisition) mode
LC-MS/MS analyses in IDA mode were performed for the unfractionated and fractionated (LAP and HAP) samples. The TOF MS survey scan was conducted in the m/z range: 100-2000 with the accumulation time of 50 ms. Top 10 precursor ions, with the charge states from +2 to +5 were chosen for the collision-induced dissociation (CID) fragmentation. Product ion spectra were collected in the m/z range of 100-2000 with the accumulation time of 40 ms. The collision energy was automatically adjusted to the particular ion using rolling collision energy function. Precursor ions were excluded from reselection for 5 s after two occurrences. The duty cycle time was 1.11 seconds. The qualitative analyses in IDA mode were performed in three biological replicates, and three technical replicates per single biological replicate.
- LC-MS/MS in SWATH (Sequential Windowed of All Theoretical Fragment Ion Spectra) mode
The SWATH-MS analyses were performed for unfractionated protein samples. The parameters of SWATH-MS1 survey scan were as follows: high sensitivity mode, the m/z range: 100-2000, accumulation time of 50 ms. The parameters in the fragmentation mode were: m/z range 400-1000, divided into 25 windows, each 25 Da wide, accumulation time of 40 ms. The collision energy for each window was determined for a +2 to +5- ions centered upon the window with a spread of 2. The duty cycle time was 1.11 seconds. The quantitative SWATH analyses were performed in three biological replicates (each protein pool was subjected to three independent digestion and desalting procedures), and three technical replicates per each biological sample.
Data analysis
- Database search: IDA-MS
IDA MS/MS spectra were searched against SwissProt database (species: Homo sapiens, version 31.07.2017) by ProteinPilot (V4.5, SCIEX) using the Paragon algorithm. The search parameters were: TripleTOF 5600+ platform, Cys alkylation - iodoacetamide, digestion – trypsin, ID Focus: allow biological modifications, Search effort-thorough ID, Confidence >10%, automatic FDR (false discovery rate). Protein was considered a true positive if the following criteria were met: at least two peptides identified per protein with confidence >95% , and the FDR value < 0,1%.
- Data analysis: SWATH-MS
SWATH-MS data were processed against the MS/MS spectra libraries extracted from SwissProt database. The ProteinPilotgroup file was loaded into MS/MS All with SWATH Acquisition MicroApp 2.01 in PeakView 2.2 (SCIEX) to automatically generate a spectral library, according to the following parameters: maximum number of proteins-10000, modified peptides-allowed, peptides shared across proteins-not allowed, a maximum of 6 peptides per protein and 6 transitions per peptide. The MS/MS spectra and chromatograms of the chosen ions (XIC) from SWATH-MS analyses were compared with the spectral libraries. Starting parametres for the peptides were: [Conf] ≥ 99, FDR < 1%, XIC width: 75 ppm, offset XIC extraction window: 10 min. Retention time calibration was performed based on 3-7 peptides, uniformly distributed according to their elution time. The peptides and transitions were manually selected for further quantitative analyses.
- Statistical analysis
The t-test analysis were performed by MarkerView software version 1.2. (SCIEX).
ELISA
The expression level of RBP4 in urine supernatant of healthy subjects, FSGS, IgAN, renal cancer and prostate cancer patients was measured using Human RBP4 ELISA kit (Fine Test, Wuhan, China) following manufacturer`s instruction. The samples were diluted 3 times with Sample Dilution Buffer included in the kit. The urine creatinine levels were assessed in parallel with Creatinine (Cr) Colorimetric Assay Kit (Sarcosine oxidase method) (Elabscience Biotechnology, TX, USA) and QuantiChrom Creatinine Assay kit, DICT-500 (BioAssay Systems, CA, USA), both provided consistent results. The samples were diluted 30 and 10 times, respectively using double-distilled water. The level of uRBP4 (ng/ml) was normalized against the averaged amount of creatinine (ng/ml).
Statistical analysis of ELISA results
The box-whisker plots were done with Microsoft Excel 2013 (version 15.0.5207.1000, Microsoft Corp.). Statistical analysis was performed using the statistical package for the social scientists (SPSS version 16, IL, USA). Normality of the data was tested using the Shapiro-Wilk test. Kruskal–Wallis test, and Mann-Whitney U tests were used for comparison of differences between groups. Pearson’s correlation coefficient was used to estimate the association of urinary RBP4 levels with the degree of proteinuria and haematuria. A p-value <0.05 was considered statistically significant.
Urinary protein identification with SWATH-MS
We performed SWATH-MS analysis of 5 urine sample pools, each composed of four specimens from FSGS, IgAN, ccRCC, chRCC and healthy subject group, respectively. The pooled urine samples of FSGS and IgAN were characterized by similar degree of proteinuria (moderate-level proteinuria), as assessed by dipstick test. ccRCC, chRCC and healthy subjects had normal urine parameters. TheSWATH-MS performed on sample pools provided quantitation for 194 (FSGS), 179 (IgAN), 271 (ccRCC), 255 (chRCC) and 275 (healthy controls) urinary proteins (Confidence Interval (CI) > 95%, FDR<1%) (Supplementary Table 2). All proteins were identified by at least 2 unique peptides. A total of 447 different proteins were identified across all groups of interest, from which 87 proteins were common for each group (Supplementary Table 3). 38 proteins were found to be unique for FSGS, including 4 components of PPAR signaling pathway which is implicated in this condition [12] (Supplementary Table 3). 23 molecules were found to be exclusively secreted into the urine of IgAN (13% of the total protein content), 37 proteins were specific for chRCC (14.5% of the total protein content), 33 for ccRCC (11% of the total protein content) and 33 for healthy controls (12% of the total protein content) (Supplementary Table 3). FSGS shared more proteins with IgAN (66%), and healthy controls (66%), than with chRCC (48%), and ccRCC (44%). The comparative, quantitative analysis of the overlapping proteomes between FSGS and the reference groups revealed that Retinol-binding protein 4 is the only molecule that clearly distinguishes FSGS from the rest of the groups (p<0.05). uRBP4 protein, the carrier of vitamin A showed the highest, significant fold-change between FSGS vs healthy controls (FCH = 9.85, p = 0.004), and between FSGS vs IgAN (FCH = 4.8, p = 0.012). In this study we did not focus on comparing the non-overlapping proteomes, as the MS managed to identify only a portion of the urine-derived proteins (the most abundant proteins). Thus, we cannot conclude that the ‘disease-specific urinary proteins’ identified in the course of the research are indeed detectable only in particular disease states.
uRBP4-ELISA
For validation, urinary levels of RBP4 were assessed with ELISA for FSGS, IgAN, ccRCC, chRCC, prostate cancer and healthy control group. The amount of uRBP4 was found to be significantly increased in FSGS in relation to IgAN (p = 0.0244), ccRCC (p = 0.0042), chRCC (p = 0.0128), prostate cancer (p = 0.02) and healthy subjects (p<0.00001) (Figure 1). The levels of uRBP4 also differed significantly between IgAN (p = 0.0076), ccRCC (p = 0.015), and chRCC (p = 0.007) versus healthy controls (Figure 1). Specific concentration cut-off value of uRBP4 i.e. ≥ 200 ng/ml allowed unequivocal discrimination of renal pathologies (41% of FSGS cases, 26% of IgAN cases, 13% ccRCC cases and 14% of chRCC cases) from prostate cancer and the normal status.
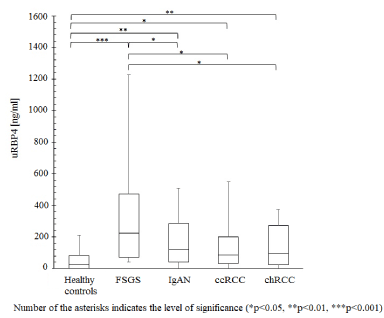
Figure 1. Concentration distribution of RBP4 in urine supernatant samples (men+women)
There was no significant relationship between the level of Retinol-binding protein 4 and the degree of proteinuria and hematuria in chRCC, ccRCC, FSGS and IgAN patients (Pearson Correlation Coefficient p > 0.05). After stratification by gender it appeared that concentration ranges of uRBP4 in glomerulopathic, renal cancer and healthy individuals are lower in women than in men (Figure 2). When only men were considered, significant differences in uRBP4 level were noted between FSGS versus healthy controls, ccRCC, chRCC, and prostate cancer patients (but not vs IgAN). In case of women, uRBP4 distribution was significantly different between healthy controls vs IgAN, ccRCC and chRCC. Because this study included only 4 FSGS females, this subgroup was omitted in the statistical analysis.
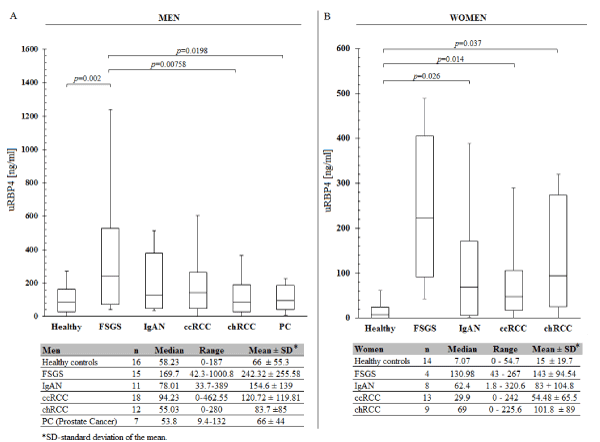
Figure 2. Concentration distribution of urinary RBP4 analysed separately for men and women
This study was initially aimed to identify urinary proteins which expression level could help distinguish FSGS patients from healthy individuals, IgAN (the second most common glomerulopathy), and renal cancer patients. Comparative analysis of the SWATH-MS protein expression profiles revealed that urinary Retinol-binding protein 4 (uRBP4), a carrier of retinol shows the highest fold change between FSGS and the rest of the groups. RBP4-ELISA performed on urine supernatant specimens from FSGS, IgAN, renal cancer, prostate cancer patients, and healthy controls (validation set) confirmed upregulation of uRBP4 in FSGS. However, it also revealed that FSGS diagnosis cannot be clearly stated based on uRBP4 concentration alone. Interestingly, it appeared that an ongoing renal disease, both glomerular disease and renal cancer can be ‘confirmed’ using specific cut-off value of uRBP4 concentration i.e. ≥ 200 ng/ml.
RBP4 is a small 21 kDa protein belonging to lipocalins, a family of proteins that facilitate the transport of small hydrophobic compounds. RBP4 is predominantly synthetized in the liver, and in lesser amount in adipose tissue (20-40%) and immune cells [13,14]. This abundant, plasma protein is mainly responsible for the redistribution of retinol (vitamin A alcohol) from the liver to peripheral tissues [15]. Possibly it also binds and transports fatty acids [16]. In circulation ̴ 86% of RBP4 remains complexed with transthyretin (TTR) as a 76 kDa unit (retinol transport complex), what prevents its leakage through the glomerular filtration barrier, and assures maintaining stable levels of retinol in the plasma [17]. The remaining 14% of the uncomplexed RBP4 is being freely filtered through the renal glomerulus, and contributes approximately 8.5 % of the proteins in the glomerular ultrafiltrate (on a molar basis) [18]. A number of studies performed in recent two decades provided evidence that uRBP4 is probably the most sensitive functional marker of the proximal renal tubule, which can detect a minor tubular damage even earlier than KIM-1 ‘biomarker’ [19-21]. Interestingly, the increase in amount of uRBP4 precedes the development of albuminuria, what indicates that uRBP4 could be potentially implemented into diagnostics and serve as a biomarker of the earliest, asymptomatic stages of renal damage. Urinary levels of RBP4 increases (even up to 104-fold) in many conditions that affect kidneys e.g. renal glomerular diseases, allograft rejection, acute kidney injury (AKI), lupus nephritis, Fanconi syndrome and diabetes [20,22-24]. Elevated levels of urinary RBP4 are also observed in tuberculosis, head and neck cancers and bladder cancer [25-27]. It has not been yet established, whether particular concentration ranges of urinary RBP4 allow identification of specific renal disorders, or help discriminate individuals with a renal/non-renal pathologies among healthy subjects.
Our study is the first report of the expression distribution of Retinol-binding protein 4 in urine samples of FSGS, ccRCC and chRCC patients, providing novel insights into the diagnostic potential of uRBP4 in renal conditions. Here we provide evidence that specific cut-off point of uRBP4 concentration ( ≥ 200 ng/ml) could be potentailly applied to detect individuals at high risk of kidney disease diagnosis (i.e. glomerulopathy, renal cancer), who should be provided with a detailed examination. We cannot exclude that the cut-off value could be lower, as the healthy controls included in our study have not undergone diagnostics for urologic diseases. The level of uRBP exceeded 100 ng/ml in only 4 from 30 healthy subjects, and in 30% individuals the protein was not or almost not detectable.
Interestingly, in our parallel project, in which we had profiled the proteomic content of urinary exosomes from FSGS, IgAN, renal cancer patients and healthy controls, no significant differences in RBP4 concentration have been observed between the groups, in fact there was a large variation in RBP4 concentration between samples from the same cohort (data not shown).
Hopefully, assessing the changes in the level of urinary RBP4 will become a detection mode of the earliest FSGS and IgAN cases (normal creatinine, nonproteinuric subjects), and the asymptomatic renal cancers (no physical signs, normal urine parameters), at the highly curable stage.
This study has several limitations i.e. no inclusion of other glomerular diseases (i.e. membranous nephropathy, minimal change disease), large variation in the degree of proteinuria and hematuria among FSGS and IgAN patients (patients included in ELISA validation study), and small sample size.
This study demonstrated that the amount of urinary Retinol-binding protein 4 is significantly raised in FSGS in comparison to IgAN, ccRCC, chRCC, prostate cancer and healthy status. Although, it shown that the expression profile of uRBP4 couldn`t make clear distinction between FSGS and the rest of diseases and the normal status, it revealed that a particular concentration cut-off value of uRBP4 could be useful in ‘recognizing’ subjects with renal pathologies (in general) among healthy individuals and prostate cancer patients. Two questions need to be addressed in prospective, validation studies: 1. Does urinary uRBP4 can be used to predict the ongoing renal disorder (in general)? 2. Which group of renal disorders can be accurately recognized by assessing the expression status of uRBP4? Further, multicenter validation studies, involving broad spectrum of patients (all types of glomerulopathies, urological cancer patients, non-urological cancer patients etc.) need to be performed to ultimately prove, whether uRBP4 possesses a diagnostic value.
Funding
This work was supported by the Leading National Research Centre (KNOW) for years 2014-2018 under Grant U17/1/4/2016; National Science Center, Sonata 12 under Grant UMO-2016/23/D/NZ5/01448.
The authors declare that they have no competing interest
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
KMB prepared samples for MS, performed ELISA experiments, analyzed and interpreted the experimental data, and was a major contributor in writing the manuscript; AK and KR collected the biological material and took part in manuscript preparation, KM and PC conducted SWATH-MS analysis and performed proteomic data analysis, ACB, PK and WW collected the biological material. All authors have read and approved the manuscript.
Not applicable.
- Stratta P, Canavese C, Marengo M, Mesiano P, Besso L, et al. (2007) Risk management of renal biopsy: 1387 cases over 30 years in a single centre. Eur J Clin Invest 37: 954-963.
- McGrogan A, Franssen CF, de Vries CS (2011) The incidence of primary glomerulonephritis worldwide: a systematic review of the literature. Nephrol Dial Transplant 26: 414-430.
- Good DM, Zürbig P, Argilés A, Bauer HW, Behrens G, et al. (2010) Naturally occurring human urinary peptides for use in diagnosis of chronic kidney disease. Mol Cell Proteomics 9: 2424-2437.
- Bidin MZ, Shah AM, Stanslas J, Seong CLT (2019) Blood and urine biomarkers in chronic kidney disease: An update. Clin Chim Acta 495: 239-250.
- Goligorsky MS, Addabbo F, O'Riordan E (2007) Diagnostic potential of urine proteome: a broken mirror of renal diseases. J Am Soc Nephrol 18: 2233-2239.
- Nafar M, Kalantari S, Samavat S, Rezaei-Tavirani M, Rutishuser D, et al. (2014) The novel diagnostic biomarkers for focal segmental glomerulosclerosis. Int J Nephrol 2014: 574261.
- Wang Y, Zheng C, Wang X, Zuo K, Liu Z (2017) Proteomic profile based screening of potential protein biomarkers in the urine of patients with nephrotic syndrome. Mol Med Rep 16: 6276-6284.
- Choi YW, Kim YG, Song MY, Moon JY, Jeong KH, et al. Potential urine proteomics biomarkers for primary nephrotic syndrome. Clin Proteomics 16: 18.
- Pérez V, López D, Boixadera E, Ibernón M, Espinal A, et al. (2017) Comparative differential proteomic analysis of minimal change disease and focal segmental glomerulosclerosis. BMC Nephrol 18: 49.
- Kalantari S, Nafar M, Rutishauser D, Samavat S, Rezaei-Tavirani M, et al. (2014) Predictive urinary biomarkers for steroid-resistant and steroid-sensitive focal segmental glomerulosclerosis using high resolution mass spectrometry and multivariate statistical analysis. BMC Nephrol 15: 141.
- Shui HA, Huang TH, Ka SM, Chen PH, Lin YF (2008) Urinary proteome and potential biomarkers associated with serial pathogenesis steps of focal segmental glomerulosclerosis. 2008. Nephrol Dial Transplant 23: 175-185
- Yang HC, Ma LJ, Ma J, Fogo AB (2006) Peroxisome proliferator-activated receptor-gamma agonist is protective in podocyte injury-associated sclerosis. Kidney Int 69: 1756-1764
- Flower DR (1996) The lipocalin protein family: structure and function. Biochem J 318: 1-14.
- Tsutsumi C, Okuno M, Tannous L, Piantedosi R, Allan M, et al. (1992) Retinoids and retinoid-binding protein expression in rat adipocytes. J Biol Chem 267: 1805-1810.
- Cho YM, Youn BS, Lee H, Lee N, Min SS, et al. (2006) Plasma retinol-binding protein-4 concentrations are elevated in human subjects with impaired glucose tolerance and type 2 diabetes. Diab Care 29: 2457-2461.
- Perduca M, Nicolis S, Mannucci B, Galliano M, Monaco HL (2018) High resolution crystal structure data of human plasma retinol-binding protein (RBP4) bound to retinol and fatty acids. Data Brief 18: 1073-1081.
- Monaco HL, Rizzi M, Coda A (1995) Structure of a complex of two plasma proteins: transthyretin and retinol-binding protein. Science 268: 1039-1041.
- Norden AGW, Burling KA, Zeni L, Unwin RJ (2019) A New Estimate of the Glomerular Sieving Coefficient for Retinol-Binding Protein 4 Suggests It Is Not Freely Filtered. Kidney Int Rep 4: 1017-1018.
- Domingos MA, Moreira SR, Gomez L, Goulart A, Lotufo PA, et al. (2016) Urinary Retinol-Binding Protein: Relationship to Renal Function and Cardiovascular Risk Factors in Chronic Kidney Disease. PLoS One 11: e0162782.
- Gonzalez-Calero L, Martin-Lorenzo M, Ramos-Barron A, Ruiz-Criado J, Maroto AS, et al. (2016) Urinary Kininogen-1 and Retinol binding protein-4 respond to Acute Kidney Injury: predictors of patient prognosis?. Sci Rep 21: 19667.
- Musial K, Zwolińska D (2018) Is urinary kim-1 the best marker of tubular damage in children with chronic kidney disease? Nephrol Dial Transplant 33: i302.
- Turnier JL, Brunner HI, Bennett M, Aleed A, Gulati G, et al. (2019) Discovery of SERPINA3 as a candidate urinary biomarker of lupus nephritis activity. Rheumatol 58: 321-330.
- Park SE, Lee NS, Park JW, Rhee EJ, Lee WY, et al. (2014) Association of urinary RBP4 with insulin resistance, inflammation, and microalbuminuria. Eur J Endocrinol 171: 443-449.
- Norden AG, Lapsley M, Unwin RJ (2014) Urine retinol-binding protein 4: a functional biomarker of the proximal renal tubule. Adv Clin Chem 63: 85-122.
- Wong SY, Cabodi M, Rolland J, Klapperich CM (2014) Evaporative concentration on a paper-based device to concentrate analytes in a biological fluid. Anal Chem 86: 11981-11985.
- Ferrari E, Wittig A, Basilico F, Rossi R, De Palma A, et al. (2019) Urinary Proteomics Profiles Are Useful for Detection of Cancer Biomarkers and Changes Induced by Therapeutic Procedures. Molecules 24: 794.
- Li F, Yu Z, Chen P, Lin G, Li T, et al. (2016) The increased excretion of urinary orosomucoid 1 as a useful biomarker for bladder cancer. Am J Cancer Res 6: 331-340.
View Supplementary Data-1
View Supplementary Data-2
View Supplementary Data-3


