Background/aim: The purpose of our study is to assess the association between the methylation level of tissue inhibitor of metalloproteinase-3 (TIMP3) and risk of renal cell carcinoma (RCC).
Methods: A systematic review of relevant literature from PubMed, EMBASE, Web of Science, and the Cochrane Library databases was conducted. Odds ratio (OR) and its corresponding 95% confidence interval (CI) were calculated to analyze the data and acquire comprehensive results.
Results: Ten eligible kinds of literature including 495 RCC specimens and 409 normal controls were analyzed in this meta-analysis. The results indicated that TIMP3 promoter hypermethylation was correlated with RCC-increased risk (OR=12.16, 95% CI: 3.64-40.66, P<0.001). The results of subgroup analysis revealed that the OR of the urine subgroup (OR=22.16) was greater than that of the tissue (OR=13.15) and blood subgroups (OR=3.20). Furthermore, the results of five studies containing RCC clinicopathological features indicated that the methylation level of TIMP3 gene was related to low tumor stage (OR=2.77, 95% CI: 1.48-5.18, P=0.001).
Conclusion: Our results suggested that the methylation level of TIMP3 promoter region was significant in RCC. Moreover, the finding is related to early tumor stage. The detection of TIMP3 promoter methylation may be a good strategy for the diagnosis of RCC at its early stages. Nevertheless, additional well-designed studies are needed to further prove our conclusion.
TIMP3, promoter, methylation, renal cancer, meta-analysis
RCC is a potentially lethal tumor, accounting for approximately 90% of all kidney cancers, and the third most frequent malignancy in the urinary system [1,2]. The incidence and mortality rates of RCC are gradually increasing at approximately 2-3% per decade worldwide [3]. In the USA, approximately 62,700 newly diagnosed subjects of kidney cancer and 14,240 deaths due to kidney cancer were reported in 2016 [4]. To date, surgical resection is the mainstream therapy for RCC and shows a good curative effect on patients with localized cancer. However, RCC in the early stage is a serious problem during diagnosis because RCC is usually asymptomatic. Approximately 20-30% of patients with RCC have already developed into distant metastasis when newly diagnosed [5]. However, no reliable biomarkers, which can support clinicians to detect RCC at an early stage, are available.
DNA methylation is one of the important epigenetic mechanisms that regulate gene expression without DNA sequence alteration and is involved in the occurrence and development of tumors [6]. Gene promoter hypermethylation accounts for most of DNA abnormal methylation and is correlated with some functional gene inactivations, particularly those involving tumor suppressor genes in human cancers [7,8]. The tissue inhibitor of metalloproteinase-3 (TIMP3) gene mapped to the chromosome 22q13.2 is a tissue inhibitor of the metalloproteinase gene family [9]. TIMP3 protein is involved in cancer progression; it disrupts the activity of vascular endothelial growth factor signaling pathway [10]. Methylation-associated silencing of TIMP3 gene plays a tumor inhibitor role in different cancers including RCC [11]. Notably, TIMP3 promoter methylation detected in urine specimens is significantly more enhanced in patients with RCC than in healthy people [12-14], indicating that it may be a noninvasive biomarker for RCC. However, the results of related studies about TIMP3 gene methylation in RCC remained controversial. In this study, we searched meticulously for relevant literature in some widely used electronic databases and conducted a meta-analysis to obtain comprehensive data.
Literature review
Related literature was identified by a systematical search in well-known electronic databases, such as PubMed, EMBASE, the Cochrane Library, and Web of Science databases, before April 2017. The following keywords were used: (methylation OR hypermethylation) AND (renal OR kidney) AND (cancer OR carcinoma OR neoplasm) AND (TIMP3 OR metalloproteinase inhibitor 3). The inclusion criteria were summarized as follows: (1) The included articles can provide sufficient information regarding TIMP3 methylation levels detected in the tissue, blood, or urine samples of patients with RCC and normal controls or regarding the estimation of the correlation between TIMP3 promoter methylation and clinicopathological characteristic of patients with RCC. (2) Studies must be published in English. (3) The RCC cases had a definite diagnosis by pathology. Recent studies or studies with large data were included only when they had repetitive data. Articles without case-control data were excluded.
Data collection
Two reviewers (S.M. and Z.S.) extracted the data independently. Useful information from each included study was extracted as follows: surname of the first author, year of publication, region, sample source, tumor histology type, sample size, tumor stage, nuclear grade, and the method of methylation detection. The specimens of normal controls were derived from healthy people or from normal tissues adjacent to RCC. Tumor stage (T1-2) was regarded as a low stage, otherwise high stage. Histological grade (G1-2) was classified as low grade, otherwise high grade.
Statistical analysis
The statistical results revealed the correlation between the TIMP3 gene methylation level and the risk of RCC. Pooled odds ratio (OR) and 95% confidence interval (CI) were used to represent the results. The Cochrane’s Q test and Higgins I2 statistic were performed to examine statistical heterogeneity [15], which was analyzed by using subgroup analysis and meta-regression. Different effect models were selected according to the results of p-value and I2 statistic. If P>0.05 or I2≦50%, a fixed-effects model (Mantel-Haenszel method) suitable for low heterogeneity was selected for use. Otherwise, a random-effects model (DelSimonian-Laird method) was selected. Furthermore, the stability of our results was evaluated by sensitivity analysis. Publication bias was inferred roughly with a funnel plot and examined quantitatively with Egger’s test and Begg’s test. All statistical data were analyzed using STATA 12.0 software (Stata Corporation, TX, USA). P<0.05 represented a significant statistical level.
Study filtration
The process of searching and selecting articles was shown in the flow diagram (Figure 1). A total of 305 kinds of literatures were identified initially by searching the databases. Then, the articles, which were duplicate or irrelevant or had insufficient data, were excluded. Eventually, 10 articles [11-14,16-21] published between 1999 and 2013 met our criteria. Among them, two articles [12,13] detected TIMP3 gene methylation not only in the tissue group but also in the urine group or blood group or both. We analyzed each group independently. The useful data of all eligible studies are listed in Table 1.
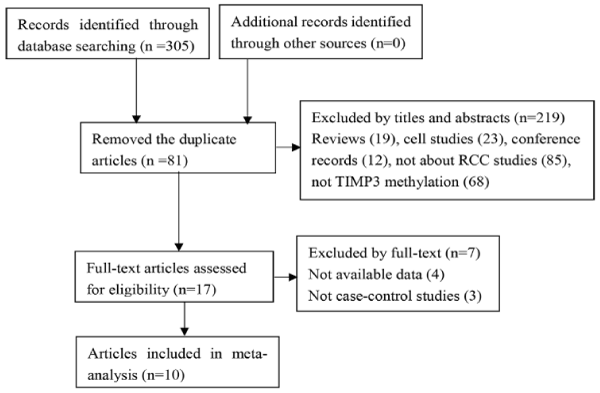
Figure 1. Flow chart of literature search and selection.
Table 1. Characteristics of the included studies. Abbreviation: RCCs: unasserted renal cell carcinoma; PC: papillary cell cancer; M: methylation; U: unmethylation; MSP: methylation-specific polymerase chain reaction; QMSP: quantitative methylation-specific polymerase chain reaction; aStudy that evaluated TIMP3 promoter methylation in urine; bStudy that evaluated TIMP3 promoter methylation in blood.
First author |
Year |
Region |
Method |
Sample type |
Histology type |
Sample size |
Case |
Control |
M |
U |
M |
U |
Bachman |
1999 |
USA |
MSP |
tissue |
RCCs |
36 |
28 |
8 |
0 |
27 |
Esteller |
2001 |
USA |
MSP |
tissue |
RCCs |
36 |
28 |
8 |
0 |
36 |
Battagli |
2003 |
USA |
MSP |
tissue |
RCCs |
45 |
26 |
19 |
0 |
10 |
Battagli-ua |
2003 |
USA |
MSP |
urine |
RCCs |
45 |
23 |
22 |
0 |
24 |
Dulaimi |
2004 |
USA |
MSP |
tissue |
RCCs |
86 |
50 |
36 |
0 |
15 |
Hoque-u |
2004 |
USA |
QMSP |
urine |
RCCs |
17 |
9 |
8 |
8 |
83 |
Hoque-bb |
2004 |
USA |
QMSP |
blood |
RCCs |
17 |
3 |
14 |
0 |
30 |
Cairns |
2004 |
USA |
MSP |
urine |
RCCs |
50 |
30 |
20 |
0 |
24 |
Costa |
2007 |
Portugal |
QMSP |
tissue |
RCCs |
75 |
13 |
62 |
15 |
47 |
Onay |
2009 |
Turkey |
MSP |
tissue |
RCCs |
21 |
4 |
17 |
2 |
19 |
Ellinger |
2010 |
Germany |
QMSP |
tissue |
PC |
32 |
2 |
30 |
0 |
15 |
Hauser |
2013 |
Germany |
QMSP |
blood |
RCCs |
35 |
20 |
15 |
21 |
33 |
Study heterogeneity
Significant heterogeneity existed in our meta-analysis (I2=80.4%). Subsequently, we proceeded with subgroup analysis and meta-regression to find the heterogeneity sources. As reported in Table 2, the data were stratified by testing method, material, and region. The detection method of quantitative methylation-specific polymerase chain reaction (qMSP) and the sample of tissue accounted for parts of heterogeneity (I2 for qMSP was 76.3% and I2 for tissue was 85.5%). In the meta-regression analysis (Table 3), heterogeneity was attributed to methods (P=0.026) but not to materials heterogeneity (P=0.607). Thus, a portion of heterogeneity was derived from the method of methylation detection.
Table 2. Subgroup analysis of the association between TIMP3 promoter methylation and RCC risk.
Variables |
Studies |
Tumor |
Control |
Pooled Effect |
Heterogeneity |
M |
U |
M |
U |
OR (95%) |
Z |
P |
I2 (%) |
P |
Total |
12 |
236 |
259 |
46 |
363 |
12.16(3.64-40.66) |
4.06 |
0.000 |
80.4% |
0.000 |
Method |
|
|
|
|
|
|
|
|
|
|
MSP |
7 |
189 |
130 |
2 |
155 |
38.30(9.02-162.64) |
4.94 |
0.000 |
51.5% |
0.056 |
QMSP |
5 |
47 |
129 |
44 |
208 |
2.93(0.83-10.34) |
1.67 |
0.094 |
76.3% |
0.002 |
Material |
|
|
|
|
|
|
|
|
|
|
Tissue |
7 |
151 |
180 |
17 |
169 |
13.15(1.53-112.72) |
2.35 |
0.019 |
85.5% |
0.000 |
Blood |
2 |
23 |
29 |
21 |
63 |
3.20(0.65- 15.67) |
1.43 |
0.020 |
33.3% |
0.221 |
Urine |
3 |
62 |
50 |
8 |
131 |
22.16(5.57-88.10) |
4.40 |
0.000 |
24.8% |
0.264 |
Region |
|
|
|
|
|
|
|
|
|
|
America |
8 |
197 |
135 |
8 |
249 |
32.33(13.32-78.45) |
7.69 |
0.000 |
8.9% |
0.361 |
Europe |
4 |
39 |
124 |
38 |
114 |
1.33( 0.63-2.79) |
0.75 |
0.456 |
29.4% |
0.236 |
Table 3. Meta-regression analysis.
Variables |
Coefficient |
95% Confidence interval |
I 2-residual |
Adjusted R2 |
P value |
Method |
-2.53 |
(-4.67,-0.38) |
65.24% |
45.93% |
0.026 |
Material |
0.38 |
(-1.21,1.96) |
75.70% |
-7.38% |
0.607 |
Region |
-3.24 |
(-4.69,-1.79) |
8.57% |
88.78% |
0.001 |
Relation between TIMP3 promoter methylation and RCC risk
Frequency of TIMP3 promoter hypermethylation in RCC cases and normal controls. As shown in forest plot (Figure 2), TIMP3 promoter hypermethylation was significantly correlated with the incidence rate of RCC (OR=12.16, 95% CI: 3.64-40.66, P<0.001, random-effects model). In subgroup analysis, the OR of urine sample subgroup (OR=22.16) was greater than that of tissue (OR=13.15) and blood sample subgroups (OR=3.20).
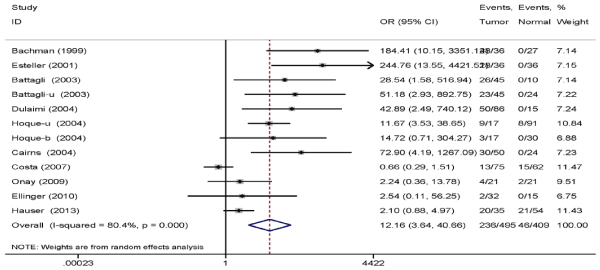
Figure 2. Pooled OR from 10 studies including 495 RCC cases and 409 normal controls. OR=12.16, 95% Cl: 3.64–40.66, P<0.001.
Association between TIMP3 promoter hypermethylation with RCC clinicopathological characteristic. The data on RCC clinicopathological features (tumor stage and histological grade) are shown in Table 4. Five studies containing definite clinicopathological information were included in the analysis, and the results indicated that TIMP3 promoter methylation was associated with low tumor stage (OR=2.77, 95% CI: 1.48-5.18, P=0.001, fixed-effects model) (Figure 3) but not associated with histological grade (OR=1.70, 95% CI: 0.98-2.95, P=0.06, fixed-effects model) (Figure 4).
Table 4. Clinicopathological characteristics of five studies. Abbreviation: M: methylation; U: unmethylation.
First author |
Year |
Method |
Sample type |
Low Grade |
High Grade |
Low stage |
High stage |
M |
U |
M |
U |
M |
U |
M |
U |
Bachman |
1999 |
MSP |
tissue |
14 |
5 |
14 |
3 |
20 |
5 |
8 |
3 |
Battagli |
2003 |
MSP |
tissue |
18 |
8 |
8 |
11 |
23 |
11 |
3 |
8 |
Battagli-u |
2003 |
MSP |
urine |
16 |
10 |
7 |
12 |
20 |
14 |
3 |
8 |
Dulaimi |
2004 |
MSP |
tissue |
29 |
18 |
21 |
18 |
39 |
22 |
11 |
14 |
Onay |
2009 |
MSP |
tissue |
4 |
15 |
0 |
2 |
4 |
14 |
0 |
3 |
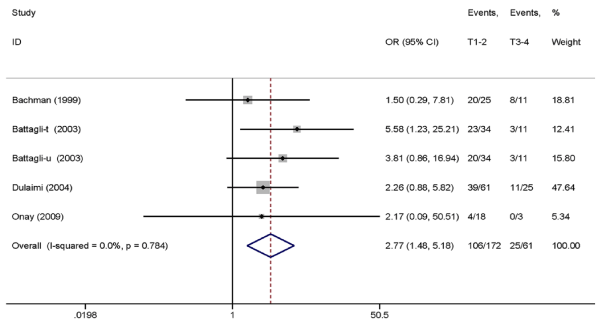
Figure 3. Pooled OR from five studies assessing the association between TIMP3 promoter methylation and tumor stage. OR=2.77, 95% CI: 1.48–5.18, P=0.001.
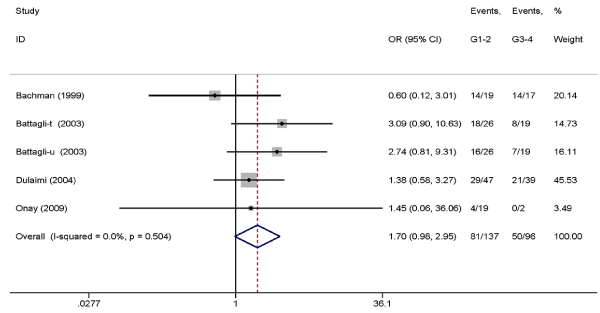
Figure 4. Pooled OR from five studies assessing the association between TIMP3 promoter methylation and histological grade. OR=1.70, 95% CI: 0.98-2.95, P=0.06.
Sensitivity analyses
Sensitivity analysis was performed to reflect the stability and credibility of the statistical results by assessing the effect of the removal of a single study on the total pooled ORs. As presented in Figure 5, the pooled ORs were not changed obviously, indicating that the results of our meta-analysis were stable.
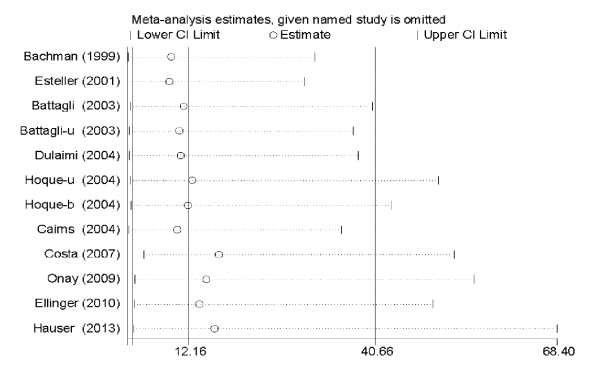
Figure 5. Sensitivity analysis of overall odds ratio coefficients for TIMP3 methylation.
Publication bias
As shown in Figure 6, the shape of funnel plot exhibited obscured symmetry, then, Egger’s and Begg’s tests were performed to evaluate publication bias quantitatively. The p-value of Egger’s test was 0.002, whereas the that of Begg’s test was 0.858. Trim and fill analysis was conducted to determine the conditions wherein the conclusions of the two tests are inconsistent. The results of the relationship between TIMP3 methylation and RCC risk also had statistical significance after increasing 5 imputed studies in trim and fill analysis (OR=1.29, 95%CI: 0.26-2.35, P=0.02). Thus, the results of Begg’s test and trim and fill analysis demonstrated that there were no significant publication bias in our meta-analysis.
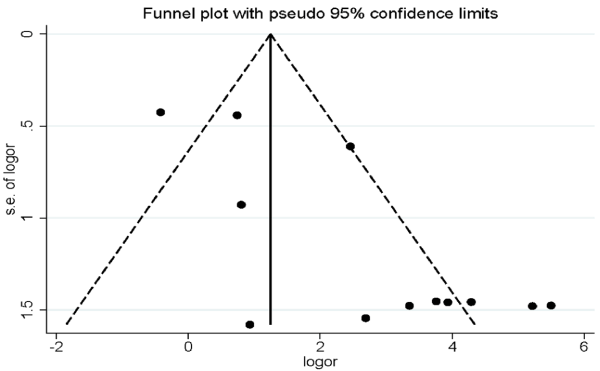
Figure 6. Funnel plot of publication biases on the correlation between TIMP3 promoter methylation and RCC risk.
DNA aberrant methylation regulates gene expression without changing DNA sequences and is extensively involved in oncogenesis. The hypermethylation of gene promoter region is associated with transcriptional silencing and inactivation of some functional genes, such as tumor suppressor genes and DNA damage repair genes in human cancers [22,23]. TIMP3 gene, found in chromosome 22q13.2, is a tumor suppressor gene. It is involved in the inhibition of the growth, angiogenesis, invasion, and metastasis of human cancers [24-26]. Decrease in TIMP3 expression is associated with abnormal promoter methylation in various malignant tumors, including kidney cancer [11,27], bladder cancer [28], choriocarcinoma [29,30], esophageal squamous cell carcinoma [31], meningioma [32], and gastric cancer [33].
However, studies on the relationship between TIMP3 promoter methylation and RCC risk have inconsistent results. During meta-analysis, TIMP3 promoter methylation was detected in tissue, blood, and urine samples through the testing method of methylation-specific polymerase chain reaction (MSP) or qMSP. The results were correlated with the increased risk of RCC (OR=12.16, P=0.000). However, distinct heterogeneity was presented in this meta-analysis. Meta-regression and subgroup analysis showed that parts of heterogeneity were derived from the method of methylation detection. Usually, the detection method of qMSP has higher sensitivity than that of MSP [34]. As previous studies reported, TIMP3 promoter methylation occurred in some normal renal tissue samples when the qMSP method was used; this result was difficult to obtained when MSP was performed [16,19]. Meanwhile, morphologically normal human tissues might undergo processes that can lead to tumorigenesis, particularly aberrant methylation because of the “field-effect” (also known as field cancerization) and gradual cytogenetic alteration [35]. The results of subgroup analysis stratified by the testing method revealed that TIMP3 promoter methylation detected only by MSP is significantly associated with RCC risk (OR=38.30). This finding suggests that our results were not influenced by the heterogeneity caused by the method of methylation detection. Basing on the results of the subgroup analysis stratified by material, we also found that TIMP3 promoter methylation detected in the urine subgroup was highly correlated with RCC-increased risk (OR=22.16). Thus, TIMP3 promoter methylation might be developed into a noninvasive and convenient biomarker to diagnose RCC.
Previous studies reported that the hypermethylation level of TIMP3 gene is associated with tumor stage [11,13,18] and histological grade [18], whereas others reported no correlation [12,16]. Five studies were conducted to produce RCC clinicopathological data to assess the relationship between the frequency of TIMP3 promoter methylation and the clinicopathological features of RCC. The results suggested that the methylation level of TIMP3 promoter region is related to low tumor stage (P=0.001). We speculated that aberrant methylation of TIMP3 gene is involved in the early stage of renal tumorigenesis. Given that some tumors are accompanied with DNA methylation alteration in clinical specimens, such as blood, urine, and ascites [36,37], research on TIMP3 promoter methylation might offer a promising and noninvasive approach for the early detection and surveillance of RCC.
A few potential limitations in our study were as follows: selection bias was inevitable as we searched only in English literature and might miss some important studies published in other languages; included articles were published several years ago (1999 to 2013), and newly published studies were suggested to be included in the analysis; other clinical data, such as cancer subtypes, age, and gender, were not presented in our meta-analysis and were not correlated with TIMP3 gene methylation.
Overall, our study indicated that TIMP3 promoter hypermethylation is closely related with the risk of RCC and correlated with low tumor stage. Additional well-designed studies with sufficient sample size may clearly confirm the role of TIMP3 promoter methylation in RCC.
The research was supported by the Region Fund of the National Natural Science Foundation of China (No. 81460386).
- Kovacs G, Akhtar M, Beckwith BJ, Bugert P, Cooper CS, et al. (1997) The Heidelberg classification of renal cell tumours. J Pathol 183: 131-133. [Crossref]
- Randall JM, Millard F, Kurzrock R (2014) Molecular aberrations, targeted therapy, and renal cell carcinoma: current state-of- the-art. Cancer Metastasis Rev 33: 1109-1124. [Crossref]
- Gupta K, Miller JD, Li JZ, Russell MW, Charbonneau C (2008) Epidemiologic and socioeconomic burden of metastatic renal cell carcinoma (mRCC): a literature review. Cancer Metastasis Rev 34: 193-205. [Crossref]
- Siegel RL, Miller KD, Jemal A (2016) Cancer statistics, 2016. CA Cancer J Clin 66: 7-30. [Crossref]
- Lam JS, Leppert JT,2021 Copyright OAT. All rights reservl approaches in the therapy of metastatic renal cell carcinoma. World J Urol 23: 202-212. [Crossref]
- Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP (1998) Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res 72: 141-196. [Crossref]
- Davis CD, Uthus EO (2004) DNA methylation, cancer susceptibility, and nutrient interactions. Exp Biol Med (Maywood) 229: 988-995. [Crossref]
- Jones PA, Baylin SB (2002) The fundamental role of epigenetic events in cancer. Nat Rev Genet 3: 415-428. [Crossref]
- Apte SS, Mattei MG, Olsen BR (1994) Cloning of the cDNA encoding human tissue inhibitor of metalloproteinases-3 (TIMP-3) and mapping of the TIMP3 gene to chromosome 22. Genomics 19: 86-90. [Crossref]
- Qi JH, Ebrahem Q, Moore N, Murphy G, Claesson-Welsh L, et al. (2003) A novel function for tissue inhibitor of metalloproteinases-3 (TIMP3): inhibition of angiogenesis by blockage of VEGF binding to VEGF receptor-2. Nat Med 9: 407-415. [Crossref]
- Bachman KE, Herman JG, Corn PG, Merlo A, Costello JF, et al. (1999) Methylation-associated Silencing of the Tissue Inhibitor of Metalloproteinase-3 Gene Suggests a Suppressor Role in Kidney, Brain, and Other Human Cancers. Cancer Res 59: 798-802. [Crossref]
- Hoque MO, Begum S, Topaloglu O, Jeronimo C, Mambo E, et al. (2004) Quantitative Detection of Promoter Hypermethylation of Multiple Genes in the Tumor, Urine, and Serum DNA of Patients with Renal Cancer. Cancer Res 64: 5511-5517. [Crossref]
- Battagli C, Uzzo RG, Dulaimi E, Ibanez de Caceres I, Krassenstein R, et al. (2003) Promoter hypermethylation of tumor suppressor genes in urine from kidney cancer patients. Cancer Res 63: 8695-8699. [Crossref]
- Cairns P (2004) Detection of promoter hypermethylation of tumor suppressor genes in urine from kidney cancer patients. Ann N Y Acad Sci 1022: 40-43. [Crossref]
- Huedo-Medina TB, Sanchez-Meca J, Marin-Martinez F, Botella J (2006) Assessing heterogeneity in meta- analysis: Q statistic or I2 index? Psychol Methods 11: 193-206. [Crossref]
- Hauser S, Zahalka T, Fechner G, Müller SC, Ellinger J (2013) Serum DNA hypermethylation in patients with kidney cancer: results of a prospective study. Anticancer Res 33: 4651-4656. [Crossref]
- Esteller M, Corn PG, Baylin SB, Herman JG (2001) A gene hypermethylation profile of human cancer. Cancer Res 61: 3225-3229. [Crossref]
- Dulaimi E, Ibanez de Caceres I, Uzzo RG, Al-Saleem T, et al. (2004) Promoter Hypermethylation Profile of Kidney Cancer. Clin Cancer Res 10: 3972-3979. [Crossref]
- Costa VL, Henrique R, Ribeiro FR, Pinto M, Oliveira J, et al. (2007) Quantitative promoter methylation analysis of multiple cancer-related genes in renal cell tumors. BMC Cancer 7: 133. [Crossref]
- Onay H, Pehlivan S, Koyuncuoglu M, Kirkali Z, Ozkinay F (2009) Multigene methylation analysis of conventional renal cell carcinoma. Urol Int 83: 107-112. [Crossref]
- Ellinger J, Holl D, Nuhn P, Kahl P, Haseke N, et al. (2011) DNA hypermethylation in papillary renal cell carcinoma. BJU Int 107: 664-669. [Crossref]
- Singal R, Ginder GD (1999) DNA methylation. Blood 93: 4059-4070. [Crossref]
- Herman JG, Baylin SB (2003) Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med 349: 2042-2054. [Crossref]
- Bodnar M, Szylberg L, Kazmierczak W, Marszalek A (2015) Tumor progression driven by pathways activating matrix metalloproteinases and their inhibitors. J Oral Pathol Med 44: 437-443. [Crossref]
- Adissu HA, McKerlie C, Di Grappa M, Waterhouse P, Xu Q, et al. (2015) Timp3 loss accelerates tumour invasion and increases prostate inflammation in a mouse model of prostate cancer. Prostate 75: 1831-1843. [Crossref]
- Han XG, Li Y, Mo HM, Li K, Lin D, et al. (2016) TIMP3 regulates osteosarcoma cell migration, invasion, and chemotherapeutic resistances. Tumour Biol 37: 8857-8867. [Crossref]
- Masson D, Rioux-Leclercq N, Fergelot P, Jouan F, Mottier S, et al. (2010) Loss of expression of TIMP3 in clear cell renal cell carcinoma. Eur J Cancer 46: 1430-1437. [Crossref]
- Hoque MO, Begum S, Brait M, Jeronimo C, Zahurak M, et al. (2008) Tissue inhibitor of metalloproteinases-3 promoter methylation is an independent prognostic factor for bladder cancer. J Urol 179: 743-747. [Crossref]
- Feng H, Cheung AN, Xue WC, Wang Y, Wang X, et al. (2004) Down-regulation and promoter methylation of tissue inhibitor of metalloproteinase 3 in choriocarcinoma. Gynecol Oncol 94: 375-382. [Crossref]
- Xue WC, Chan KY, Feng HC, Chiu PM, Ngan HY, et al. (2004) Promoter hypermethylation of multiple genes in hydatidiform mole and choriocarcinoma. J Mol Diagn 6: 326-334. [Crossref]
- Ninomiya I, Kawakami K, Fushida S, Fujimura T, Funaki H, et al. (2008) Quantitative detection of TIMP-3 promoter hypermethylation and its prognostic significance in esophageal squamous cell carcinoma. Oncol Rep 20: 1489-1495. [Crossref]
- Barski D, Wolter M, Reifenberger G, Riemenschneider MJ (2010) Hypermethylation and transcriptional downregulation of the TIMP3 gene is associated with allelic loss on 22q12.3 and malignancy in meningiomas. Brain Pathol 20: 623-631. [Crossref]
- Guan Z, Zhang J, Song S, Dai D (2013) Promoter methylation and expression of TIMP3 gene in gastric cancer. Diagn Pathol 8: 110. [Crossref]
- Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, et al. (2000) MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res 28: E32. [Crossref]
- Chai H, Brown RE (2009) Field effect in cancer-an update. Ann Clin Lab Sci 39: 331-337. [Crossref]
- Bryzgunova OE, Morozkin ES, Yarmoschuk SV, Vlassov VV, Laktionov PP (2008) Methylation-specific sequencing of GSTP1 gene promoter in circulating/extracellular DNA from blood and urine of healthy donors and prostate cancer patients. Ann N Y Acad Sci 1137: 222-225. [Crossref]
- Jung M, Putzer S, Gevensleben H, Meller S, Kristiansen G, et al. (2016) Diagnostic and prognostic value of SHOX2 and SEPT9 DNA methylation and cytology in benign, paramalignant, and malignant ascites. Clin Epigenetics 8: 24. [Crossref]






