Abstract
A pair of vertebrate-specific and brain-expressed pre-synaptic genes, NTNG1 and NTNG2, contributes to the Intelligence Quotient (IQ) test scores in a complementary manner. Single nucleotide polymorphisms (SNPs) of NTNG1 are associated with attenuated verbal comprehension (VC) or processing speed (PS) while NTNG2 SNPs affect working memory (WM) and perceptual organization (PO), forming cognitive endophenotypes in healthy and schizophrenia (SCZ)-affected human subjects. Regions of interest (ROIs), defined as 21 nucleotide (nu) long NTNG gene loci symmetrically embedding the IQ-affecting mutation alleles (VC/PS and WM/PO), underwent dramatic evolutionary changes from mice through primates to hominins, at the accelerated rates. Mutation alleles associated with the higher VC and WM IQ scores are found in the genomes of extinct hominins of Neolithic times, however, lower WM scores associated allele is also detectable in Mesolithic hunters genomes. Protein sequence of NTNG1 is 100% conserved among the primates, archaic and modern extinct hominins while NTNG2 underwent a recent selection sweep encoding a primate-specific S371A/V (~50,000 yrs BC), and a modern human (5,300 yrs BC) T346A substitutions. We show that a 500 mln yrs old genomic duplication of a synapse primordial gene provided a substrate for further synapse elaborations and its ultimate capacitive expansion of what evolved into a vertebrate cognitive superior complexity – intelligence.
Key words
evolution, gene duplication, psychogenetics, cognitive endophenotype, paleogenetics
Introduction
S.Ohno ingeniously proposed that new gene function can result from gene duplication and following it gene paralogs subfunctionalisation (SF) [1]. Two gene paralogs, NTNG1 and NTNG2, expressed predominantly in the brain [2], and encoding Netrin-G1 and Netrin-G2 proteins, respectively, are expressed pre-synaptically and segregate in a non-overlapping manner into distinct neuronal circuits [3,4]. They are related to the netrin family of axonal guidance cues [5] but differ in that they attach to the axonal membrane via a glycosylphosphatidylinositol (GPI) link [6,7], a known lipid raft-associated membrane signaling cascade organiser [6,8]. The first evolutionary precursor of NTNG as a single gene copy can be located in the genome of a primitive vertebrate tunicate/urochordate Ciona intestinalis (sea squirt, ENSCING00000024925), reported to be the first organism with the neural crest primordials [9], multipotent brain progenitor cells [10], and neurogenic placodes, facilitating the transition from pelagic invertebrate life style to a predatory vertebrate [11]. The dramatic expansion of human cerebral cortex over the course of evolution [12-15] had provided new niches for accommodating either de novo or advancing pre-existed cognitive features and culminating in the positively selected human cognitive functions [16].
IQ tests are a surrogate measure of general human cognitive ability characterising intelligence. They are often administered as WAIS-III/IV [17] and represent a cumulative score of 4 cognitive indices: VC (verbal comprehension), WM (working memory), PO (perceptual organization) and PS (processing speed) frequently referred as “cognitive domains” [18]. IQ has been validated by factor analyses [19], and a common factor (correlate) influencing each of them referred as g, or “general intelligence”, proposed by Spearman [20], but recently challenged as the only existing correlate [21]. IQ is affected by several mental disorders including schizophrenia (SCZ) characterised by severe cognitive deficits in WM [22], behavioral flexibility and resulting in low performance in cognitive tests [23-25]. An existence of quantitative trait loci associated with WM endophenotype and SCZ diagnosis has been predicted before [26]. Since both NTNG paralogs have been reportedly associated with SCZ [27-35] we investigated whether these gene paralogs contribute to human intelligence by assessing the IQ of human carriers for the NTNG1 and NTNG2 SNP alleles against non-carriers with and without SCZ.
Results
Several NTNG1 and NTNG2 SNPs have been previously reported to be associated with SCZ (e.g. 28, 31). We have found that out of 11 SNPs tested, five affect the IQ scores and composite domains in human subjects (Figure 1, Supplementary Table 1 (ST1)). SCZ patients carrying rs2218404 T allele (Figure 1A-1) of NTNG1 (T/G and T/T genotypes, N = 25 patients) compared with G/G genotype (N = 36 patients) demonstrated attenuated full-scale IQ (FIQ, ANCOVA p = 0.0057 (F1,198 = 7.80, partial η2 = 0.038)), and verbal IQ (VIQ) (p = 0.0033 (F1,198 = 8.87, partial η2 = 0.043)). Verbal comprehension (VC) domain score was the main contributor to the VIQ decline (p = 0.0050 (F1,198 = 8.08, partial η2 = 0.039), Figure 1B-1), with low scores across all parameters except comprehension (CH): vocabulary (Vc, p = 0.020 (F1,198 = 5.49, partial η2 = 0.027)), similarities (SiM, p = 0.041 (F1,198 = 4.23, partial η2 = 0.021)), and information (IF, p = 0.0067 (F1,198 = 7.50, partial η2 = 0.037), Figure 1B-1 lower panel). Thus, a mutation allele of NTNG1, rs2218404, is associated with low VC and affects the VIQ via the attenuated Vc, SiM and IF subscores. The next NTNG1 SNP found to affect IQ was rs96501, with attenuated processing speed (PS) in healthy human subjects C allele carriers (N = 45) vs T/T genotypes (N = 98, p = 0.028 (F1,190 = 4.89, partial η2 = 0.025), Figure 1B-1) with no effect on SCZ patients. The contributing affecting score was symbol search (SS, p = 0.053 (F1,190 = 3.79, partial η2 = 0.020), Figure 1B-1, lower panel) with digit symbol coding (DSC) being also attenuated but non-significantly (p = 0.12 (F1,198 = 2.40, partial η2 = 0.012)). Three other SNPs mapped to NTNG2 (Figure 1A-2) have been also shown to affect IQ. Healthy carriers of the NTNG2 SNP rs1105684 A allele (N = 49) showed a lower FIQ (p = 0.018 (F1,141 = 5.70, partial η2 = 0.039)), VIQ (p = 0.029 (F1, 141 = 4.90, partial η2 = 0.034)), and performance IQ (PIQ) (p = 0.048 (F1, 141 = 3.99, partial η2 = 0.028)) when compared with the T/T genotypes (N = 96, Figure 1B-2). To check for a potential dosage-dependent effect of a mutation allele on IQ score, NTNG2 SNP rs2149171 SCZ and healthy human subject cohorts were each split on 3 genotypes, respectively: C/C (N = 14 and 39), C/T (N = 29 and 73), and T/T (N = 15 and 30). The presence of the C allele as a single copy (C/T genotype) was strongly associated with a prominent attenuation in the IQ scores of SCZ patients and was essentially identical to that produced by the C/C genotype when both are compared to the T allele carriers (FIQ: p = 0.014 (F2,192 = 4.35, partial η2 = 0.043); VIQ: p = 0.029 (F2,192 = 3.60, partial η2 = 0.036); PIQ: p = 0.035 (F2,192 = 3.42, partial η2 = 0.034), Figure 1B-2). If in the case of NTNG1 located SNP rs2218404 the lower VIQ score was contributed mainly by the decreased VC domain scores for Vc, SiM, and IF (Figure 1B-1), in the case of NTNG2 located rs2149171 the CH and WM domain scores were responsible for the VIQ decline in C allele carriers (p = 0.012 (F2,192 = 4.54, partial η2 = 0.045) for CH and p = 0.040 (F2,185 = 3.27, partial η2 = 0.034) for WM (N = 12 (C/C); N = 25 (C/T) and N = 14 (T/T)). Similarly to VIQ, where CH of rs2149171 complements the cognitive endophenotype produced by the T-allele of rs2218404, the PIQ attenuated score in the case of rs2149171 was due to lower PO score (p = 0.050 (F2,192 = 3.04, partial η2 = 0.031), Figure 1B-2) in the C allele carriers. The third NTNG2 located SNP found to affect the human IQ was rs2274855 (Figure 1A-2) with a cognitive endophenotype associated with the A-allele presence in SCZ patients (N = 33) vs. G/G genotypes (N = 26) and resembling that of the described above C-allele of rs2149171. Accordingly, the attenuated scores were: FIQ (p = 0.012 (F1,196 = 6.44, partial η2 = 0.032)), VIQ (p = 0.017 (F1,196 = 5.83, partial η2 = 0.029)), and PIQ (p = 0.036 (F1,196 = 4.46, partial η2 = 0.022), Figure 1B-2). Similarly to rs2149171 the lower VIQ score was due to declined CH (p = 0.035 (F1,196 = 4.49, partial η2 = 0.022)) but unaffected VC that is contrary (complementary) to the rs2218404 endophenotype (Figure 1B-1). WM was robustly affected by the A allele presence (N = 29; p = 0.023 (F1,189 = 5.28, partial η2 = 0.027)) contributed by the low digit span (DS) score (p = 0.026 (F1,196 = 5.04, partial η2 = 0.025)) with letter number sequencing (LNS) and arithmetic (AM) being unaffected (Figure 1B-2, lower panel). The observed PO score was comprised by the declined matrix reasoning (MR, p = 0.024 (F1,196 = 5.19, partial η2 = 0.026)), block design (BD, p = 0.010 (F1,196 = 6.82, partial η2 = 0.034)) with picture completion PC being unchanged (Figure 1B-2, lower panel). Thus, all three aforementioned NTNG2 SNPs affect the VIQ and PIQ in human subjects with the first one contributed by the CH subscore and WM and the latter by the PO score (Figure 1C). Contrary to this, the NTNG1 located SNP (rs2218404) affects the VIQ through the lower Vc, SiM and IF scores and affecting the VC domain scores. Another SNP, rs96501, affects PS domain, although in healthy subjects only. It can be concluded that both genes (as paralogs) contribute to the cognitive scoring produced upon the implemented IQ testing but in a cognitive domain-complementary manner. NTNG1 contributes to the VC and PS while NTNG2 for the WM and PO domain scores (Figure 1C).
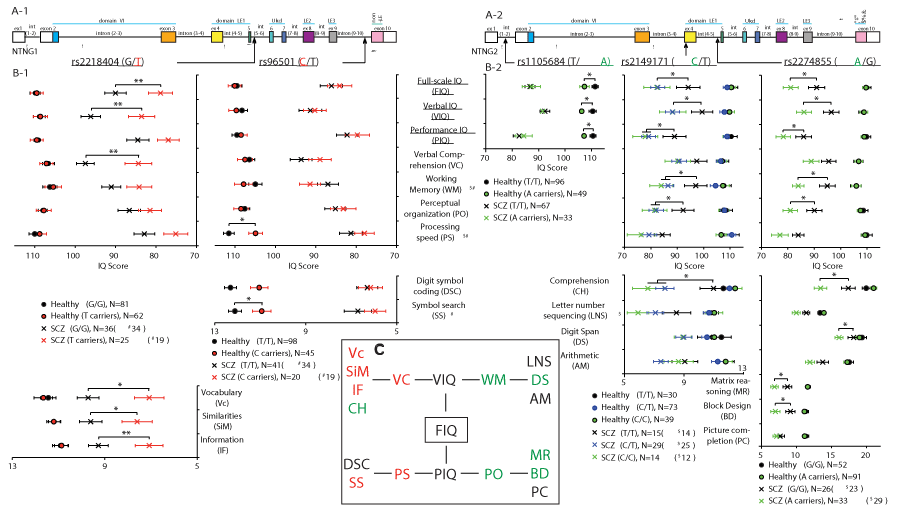
Figure 1. Complementary effect of NTNG paralog SNPs on IQ cognitive domains of human subjects as measured by WAIS-III. (A-1, A-2) NTNG1 and NTNG2 gene structures with the SNPs location indicated. (B-1, B-2, C) Affected cognitive domains and (sub)scores. Red highlights NTNG1 and green (or blue, in case of heterozygosity) NTNG2-located alleles associated with the attenuated IQ scores. The data are presented as a mean+SEM. *p<0.05,**p<0.01 (two ways ANCOVA (sex, education, and age as co-variates)). The number of human subjects is indicated as N. For statistical and diagnosis details refer to ST1.
The robust link observed between a single SNP and affected cognitive domain IQ score (Figure 1) can be explained by some global dramatic perturbations caused by the presence of a mutated allele and/or a functinal importance of its context-dependent positioning on the gene (Figure 2A and 3A). To determine a potential significance of the SNP alleles’ epistatic environment we compared the nucleotide (nu) sequence within the immediate vicinity of a SNP allele positioning (50 nu upstream and downstream) in mice, primates and modern human. We compared all 11 SNPs used for the IQ screening and plotted the identity percent as a function of distance from the mutated allele position (Figure 2B and 3B and Supplementary Materials). We found that the identity percent distribution over the analysed areas of ±50 nu is not uniform and displays a SNP allele position-centred dramatic evolutionary changes pointing towards a potential functional significance of the immediate vicinity of a SNP as short as ±10 nu and not further, referred from here and beyond as a Region Of Interest (ROI) for each specific SNP allele. We calculated the rates of evolutionary changes for each ROI as a percent identity change over the lapsed mln yrs of evolution (Figure 2C and 3C). Among the 6 NTNG1-located SNP ROIs three of them display accelerated rates of evolution from marmoset to chimpanzee (rs2218404, rs628117, rs96501) when compared to the mouse-marmoset rates, and rs2218404 (affecting VC in human subjects) additionally demonstrates an accelerated rate of evolution on the chimpanzee to human path (Figure 2C). As for the NTNG2 located ROIs (Figure 3B), rs1105684 is remarkably consistent at displaying high evolutionary rates around 0.8 and, together with rs2274855, both have identical rates at the mouse-marmoset and chimpanzee-human paths, but differ dramatically at the marmoset-chimpanzee point (0.8 vs 0, respectively). rs2274855 is the only NTNG2-nested SNP ROI that underwent an accelerated evolution (AE) from chimpanzee to human. Next we compared the DNA sequences of all 11 ROIs across mice, primates and extinct hominins NTNG gene paralogs (see ST2 for the datasets sources used for the genes reconstruction). T-allele of rs2218404 is detectable in marmoset and its position in mouse corresponds to adenosine (Figure 2D). G-alelle (associated with a higher VC score comparing to the human T-allele carriers) is found in Mesolithic hunter Loschbour (8,000 BC) but not in another ancient hunter Motala12 and is also present in other two hominins belonging to the Neolithic period, Iceman and Eskimos (5,300 and 4,000 yrs BC, respectively). rs628117 is the only NTNG1-related mutation near vicinity of which (±50 nu) is located an intra-hominins (Es, Ice, Lo) mutation (Figure 2B, low left). The next, PS-affecting, ROI of rs96501 displays an intricate path of T-allele evolution (associated with a higher PS score) being anciently conserved from mice to primates but later substituted on the less efficient (in terms of the generated IQ scores) C allele in Neanderthals, later again replaced by the T allele in Mesolithic hunters and coming back during the Neolithic times (Figure 2D). The first NTNG2 SNP rs1105684 is located at the beginning of the gene and affects WM in healthy human subjects (Figure 3A). The origin of the T-allele is evolutionary bound to marmoset since its position in mice is occupied by another pyrimidine base C (Figure 3D). Next on the gene are two SNP alleles for rs7851893 and rs3824574 which do not affect IQ and similarly to rs1105684 are surrounded by highly conserved ROIs not only in hominins and chimpansee (100% identity) but also in marmoset (except 1 mutation for rs7851893). rs2149171 ROI (affecting the WM and PO scores) similarly to rs3824574 is 100% conserved across the all species (including the mouse) except the allele itself. The attenuating IQ C-allele position is occupied in mice genome by T but present in Iceman and Eskimos genes. A distinct evolutionary path is taken by another cognitive endophenotype-associated and affecting WM and PO scores A-allele of rs2274855 and its ROI (Figure 3D). Its position in mice is likely to be occupied by the C pyrimidine base (the software places a blank instead of it) which is gradually substituted on purine G in chimpanzee and misplaced by the lower IQ score-associated A-allele in Mesolithic hominins. And 20 nu downstream of the centre of ROI is located a modern human-specific point mutation translated into the T346A protein substitution (as described below).
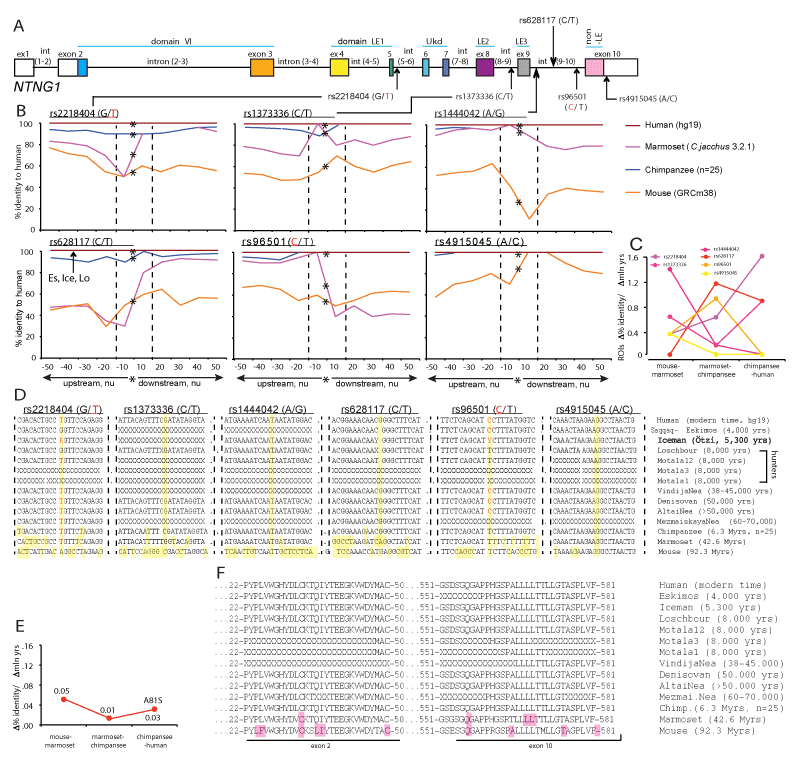
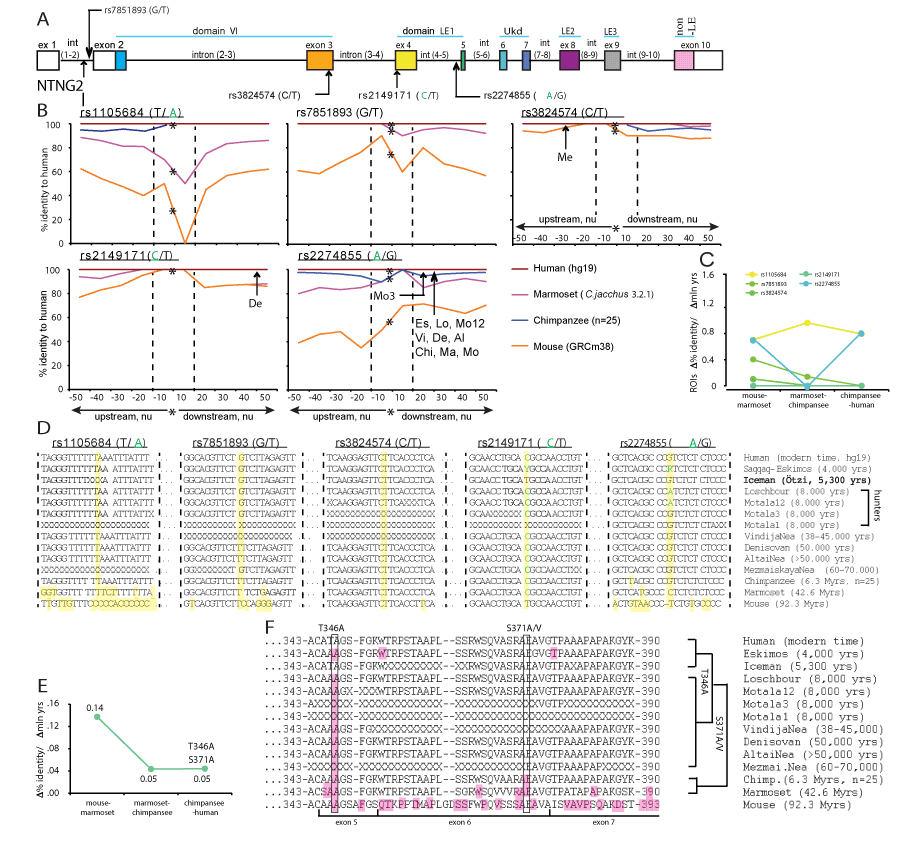
Figures 2 and 3. Accelerated evolution (AE) and definition of ROI of NTNG loci embedding associated with the cognitive endophenotype alleles. Hominins and primate-specific protein amino acid substitutions. (A) NTNG1 and NTNG2 SNPs’ gene locations. Alleles associated with the lower IQ scores are shown either in red (NTNG1) or green (NTNG2). (B) Calculated identity percent of primates and mice to human gene loci as a function of distance from the position of a mutated allele (denoted as a star). Comparison was done in a stepwise manner as ±10 nu to the maximum of ±50 nu using Stretcher (http://www.ebi.ac.uk/Tools/psa [71]). The areas were compared based on the positioning relative to the point mutation without any manual curation for “the best-fit” alignment, but as per the algorithm output only. Due to low level of identity for mouse and marmoset the initial search of the mutation allele positioning was done aligning against the human corresponding full-length intron, and then second time against the obtained 101 nu query (“-50 nu-SNP+50 nu”). Two dashed lines define an area of -10 nu to +10 nu from the mutation allele position. This area (21 nu in total) is defined as a ROI of the given mutation allele of a representing SNP. An arrow indicates a position of extinct hominins-specific mutations (see below). (C) Evolutionary rates for the ROIs calculated as a percent identity change relative to the hg19 over the mln of years of evolution. The spectrum color reflects the mutations’ positioning order on a gene, as purple-yellow for NTNG1, and yellow-blue for NTNG2. (D) ROIs DNA sequences across hominins, primates and mice loci. The extinct and ancient hominin’s NTNG paralogs were reconstructed from the available datasets (see SM): Saqqaq-Eskimos (Es) [60]; Iceman (Ice) [59]; Hunters (Loschbour (Lo), Motala12 (Mo12), Motala3 (Mo3), Motala1 [58]; VindijaNea (Vi) [73]; Denisovan (De), AltaiNea (Al) and MezmaiskayaNea (Me) [74]; chimpanzee (Chi), reconstructed from [72], n = 25 animals). Non-readable sequences due to poor reads quality are denoted as X. Ensemble was used for the initial NTNG paralogs retrieval in marmoset (Ma) and mouse (Mo). Yellow (vertical strip) denotes the position of the SNP-related mutation allele and non-matched to human substitutions. (E) Evolution rates for the proteins encoded by the NTNG1 (Netrin-G1) and NTNG2 (Netrin-G2). (F) Amino acid changes for Netrin-Gs across hominins, primates and mice. For Netrin-G1, there are no common mutations among primates, hominins and modern human. Es contains 5 point mutations absent in other hominins; Chi contains 1 mutation outside the depicted area (A81S); all 4 mutations for marmoset are shown, and 10 more extra mutations for mouse are not shown. For Netrin-G2, all hominins except Ice (relative to hg19) carry a T346A mutation (exon 5-located and known as rs4962173), also detectable in primates and mouse. Chi’s Netrin-G2 differs from all hominins by S371A mutation (exon 6 - nested) and present in marmoset as S371V. Non-matched to hg19 amino acids are highlighted as pink. Refer to SM for the full alignments.
The distinct picture of evolutionary changes among the NTNG1 and NTNG2 nested SNPs has prompted us to compare evolutionary rates for the full-length proteins encoded by these gene paralogs, Netrin-G1 and Netrin-G2, respectively (2,36). Netrin-G1 undergoes only few changes in its amino acid composition with the maximum calculated rate of evolution reaching 0.05 when mice and marmoset proteins are compared, 0.01 among the primates, and 0.03 between chimpanzee and human due to a single point mutation A81S (Figure 2E and SM: Netrin-G1), absent in other primates. Netrin-G2 evolves 2.8 times faster between mouse and marmoset than its paralog, Netrin-G1, and continues evolving with a steady rate of 0.05 from primates to human (Figure 3E and SM: Netrin-G2). We have also reconstructed both proteins from ancient (Neanderthals, Paleolithic time) and extinct hominins (Mesolithic and Neolithic times) and compared them with primates’ and mice’ Netrin-G orthologs (Figure 2F and 3F). Netrin-G1 is a highly conserved protein among the primates and hominins (Figure 3F). As for Netrin-G2, a mutation shared among the Neanderthals’ and Mesolithic genomes, primates and mice (T346A) is absent in the Neolithic Iceman and modern human (the signal for Motala3, Motala1 and MezmayskayaNea is not clear due to low sequence coverage). Primates (marmoset and chimpanzee) share another mutation (S371A/V) preserved in mouse and absent in hominins (further details can be found in the SM: Results).
Discussion
NTNG paralog SNPs and associated cognitive endophenotypes of human subjects
Shortcomings of cognitive and information processing are key features of SCZ diagnosis [37]. They are frequently manifested as impairments in PO, WM, VC and PS [38] and reported as attenuated scores upon IQ tests implementation. SCZ patients carrying a mutation allele for one of NTNG gene paralog SNPs form cognitive endophenotypes affecting the IQ scores (Figure 1C). The term “endophenotype” was originally coined by John and Lewis [39] and later advanced through the field of psychiatry by Gottesman and Shields (reviewed in [40]) as a biomarker associated with a specific phenotypic trait [41]. The formed SCZ endophenotypic groups comprise from subjects with either affected VIQ (via attenuated VC by NTNG1 rs2218404 or WM by NTNG2 rs2149171 and rs2274855) or affected PIQ (via attenuated PO by NTNG2 rs2149171 and rs2274855). In two extra cases PS is affected by rs96501 of NTNG1 and WM by rs1105684 of NTNG2 (Figure 1B-1 and B-2, respectively) but in healthy human subjects. Such intriguing non-overlapping effect on the IQ domains prompts us to conclude that NTNG paralogs complement each other function and represent an example synapse-expressed genes affecting the human cognitive abilities, perhaps through the precision of neuronal connectivity perturbations and concomitant miswirings. The observed phenomena of the affected WM is the most striking due to its multifaceted constructive nature [42] underlying many, if not all, cognitive tasks such as comprehension, reasoning and learning [43] and historically introduced by Baddeley as the reading span test [44]. Lack of the localization effect of NTNG2 SNP mutation alleles, all three are located in different parts of the gene (Figure 1A-2) but associated with identical endophenotype (Figure 1B-2), points to a uniform nature of the NTNG2 function distribution over the entire gene. An obviously non-coding nature of all five IQ-affecting alleles (rs2149171 despite being exon 4-located encodes a silent F246F mutation) corroborates an idea that anthropoid trait-associated loci lie outside coding protein areas [45,46]. This hints towards a potential of these alleles to perturb genes regulatory functions, e.g. mRNA splicing, affecting downstream located pivotally functional NTNG elements such as Ukd-domain encoding exons 6 and 7 or a unique Netrin-Gs trait – GPI-link. Alternatively, or simultaneously, the NTNG SNP alleles may be embedded into an epistatic network of other genes influencing human cognitive traits [47]. However since it is usual for a SNP effect to be estimated using an additive model (assuming either independent and cumulative single contribution) to the mean of a trait with the small effect size the power to detect the epistatic environment drastically declines. Contrary to the genetic associations with gene expression having large effect sizes [47], cognitive trait-associated effect sizes are reportedly small [48], e.g. the largest effect sizes of the variance of intelligence scores accounted for only 0.2% [49], 0.5% on GWA studies of 1,583 adolescence [50] or was predicted to be ~1% on 3,511 adults [51]. Another GWAS of educational attainment (sharing a moderate correlate with intelligence), which included 126,559 individuals, reports on just 1% of the variance but only 0.02% in a replication sample [52]. Our data support the preexisted conclusion that human cognitive traits modalities are not described by statistically large effect sizes.
Evolutionary elaborations of the embedding IQ-affecting mutations loci
Eleven previously published SCZ-associated SNPs were tested for their effect on IQ performance of human subjects and 5 of them were found to be associated with attenuated IQ cognitive endophenotypes (Figure 1B-1 and B-2). ROIs of 3 of them (rs2218404, rs1373336 and rs2274855) underwent an AE from chimpanzee to human when compared to New World monkeys to apes (marmoset-chimpanzee) path (Figure 2C and 3C). Two of them affect IQ in humans (rs2218404 – VC and rs2274855 – WM, Figure 1B-1 and B-2) while being located within the vicinity of exon 5 (2,275 nu downstream and 15 nu upstream, respectively) – a part of the lowest percent identity coding DNA among the NTNG gene paralogs [53]. Presence of the evolutionary accelerated regions within the NTNG genes non-coding areas underscores them as contributors to the human-specific traits along with other genes [54]. However, not only ROIs of the IQ-affecting alleles but the alleles themselves demonstrate several unique evolutionary features (SM: Discussion). To understand evolutionary forces driving the emergence of cognitive endophenotype-associated alleles we have deduced a set of rules outlined as follows. 1. An alternative (mutated) allele evolutionary appearance coincides with the lack of any other mutations within ROI (a conserved island rule); 2. positioning of the future mutation often represents a turning point of dramatic changes of an allele ROI (e.g. as seen in marmoset: rs2218404 (50-90%), rs628117 (30-80%), rs96501 (100-40%)); 3. an AE of a ROI often precedes the emergence of a mutation allele (e.g. rs2218404: chimpanzee to human (k = 1.59), appearance of “G” in Loschbour; rs628117: marmoset to chimpanzee (k = 1.10), appearance of “T” in AltaiNea; rs96501: marmoset to chimpanzee (k = 0.83), appearance of “C” in AltaiNea; rs2274855: chimpanzee to human (k = 0.79), appearance of “A” in Motala12; 4. low identity percent (equivalent to subsequent substantial evolutionary changes) among the evolutionary species within the allele surrounding proximity of ±50 nu is not sufficient for the future mutated allele significance as a cognitive endophenotype determinant (as deduced by the IQ score) as seen for rs1373336, rs1444042, rs4915045 and rs7851893 (none of them are IQ-affecting, though associated with SCZ, despite showing (very) low identity in mice). Rather some dramatic changes within the allele’s immediate proximity of ±10 nu (defined as a ROI) preceded or followed by more stringently conserved DNA are necessary (the conserved island perturbation rule). Currently we are unable to state that the IQ-associated alleles ROIs represent regulatory loci and an important source of evolutionary innovation [55] but they may be the smallest functional blocks of a strong positive selection that exerts its action upon, similarly to the 20–30 nu clusters of strongly conserved non-coding elements (CNEs), transcription factor binding sites (TFBS), RNA splicing and editing motifs [56].
Extinct hominins and IQ-associated mutation alleles
Availability of archaic genomes allows excavation for the advantageous alleles that modern humans acquired from archaic extinct hominins such as Neandertals and Denisovans who used to live 230,000-30,000 years ago (Middle/Upper Paleolithic, Old Stone Age) defined by distinct morphological features [57], and from modern extinct humans (hunters, farmers) from Mesolithic (Middle Stone Age, ~10,000 yrs BC, [58]) and Neolithic (New Stone Age, ~5,000 BC, [59,60] periods. Though exhibiting several anatomical features, making archaic hominins different from the modern human, there are studies challenging the idea that reserve symbolism and abstract thinking was an exclusive prerogative of modern human [61,62]. The time Neanderthals used to live in is thought to be associated with the onset of cognitive fluidity involving the capacity to draw analogies (early paintings), to combine concepts (making tools) and to adapt ideas for new contexts [63]. Wynn and Coolidge believe that evolution of WM was central to the evolution of human cognitive traits consisted from few genetic mutations that led to “enhanced WM” 200,000-40,000 BC [64]. Our work partially supports this idea showing the perseverance of higher WM score-associated alleles across the hominins such as T of rs1105684 and G of rs2274855 (Figure 3D) but a Neolithic appearance of rs2149171 T in Iceman previously found only in mice genome supporting the conclusion made by Crabtree [65] that modern humans as species “are surprisingly intellectually fragile and perhaps reached a peak 2,000–6,000 years ago”. Mesolithic period has been always considered as a key gate for the evolution of human languages [66] with our data showing that rs2218404 G allele associated with a higher VC score (Figure 1B-1) emerges for the first time in the Loschbour hunter NTNG1 gene (Figure 3D). Verbal comprehension, as an example of abstract symbol usage, is associated with the global network efficiency (as a part of fronto-parietal network [19,67], and global communication and intellectual performance [68]. From this point of view it is not surprising that appearance of the G allele in Loschbour and Iceman genomes coincides with the presence of PS enhancing rs96501 T allele (Figure 3D). A wealth of data has been collected characterising possible look and health status of archaic hominins and modern but extinct humans [69] (SM: Discussion). Based on our own data we may also speculate that the extinct hominins may have had a lower VC comparing to us, and consequently, Neanderthals were unlikely able for a semantic communication due to a global network inefficiency VC is associated with; they have had similar to us PS (if they had lived beyond the Mesolithic period), and were likely to have had identical to modern human WM, corroborating the advanced evolutionary nature of this important human cognitive domain of a limited capacity and associated with intelligence.
Conclusion
Evolution of a novel function relies on enhanced genetic robustness via functional redundancy potentially provided by a gene duplication event. Further evolutionary outcome depends on the substrate availability (undergoing its own evolution) upon which the novel function(s) exerts its action. Nature does not create but tinkers to perfection provided to it material exploring available evolutionary tools. Half a billion years ago a gene duplication event had provided a plethora of such substrate thus converting the evolution itself into a “Creator” of new functions. Here we have described how a pair of twin genes got themselves involved into the human cognitive functioning believed to emerged in a primordial state in primitive vertebrates prior to the first recorded gene duplication. Subsequent process of the function specialisation made NTNG paralogs to subfunctionalise into distinct cognitive domains in a complementary manner [53].
Materials and methods
Ethics statement
This study was performed in accordance with the World Medical Association’s Declaration of Helsinki and approved by the Osaka University Research Ethics Committee. A written informed consent was obtained from all subjects after the procedures had been fully explained.
Subjects
The procedures were performed as per established protocols at Osaka University as described previously [70]. The subjects consisted from 100 patients with SCZ and 145 healthy controls. The sex ratio and the mean age did not differ significantly between the groups. The subjects were all biologically unrelated Japanese and recruited from both outpatient and inpatient units at Osaka University Hospital and other psychiatric hospitals. Each patient with SCZ had been diagnosed by at least two trained psychiatrists based on unstructured clinical interviews, according to the criteria of the DSM-IV [37]. In case if the diagnosis of the two trained psychiatrists was discordant, it was resolved through the further negotiations on both specialist opinions. In case of unresolved diagnostic disputes, the patient was omitted from the study. Psychiatrically healthy controls were recruited through local advertisements and were evaluated by means of unstructured interviews to exclude individuals with current or past contact with psychiatric services, those who experienced psychiatric medications, or who were not Japanese. Controls for family history of a cognitive disorder, such as SCZ, bipolar disorder, or major depressive disorder were not included. Ethnicity was determined by self-report and was not confirmed by genetic analyses. Additionally, subjects were excluded from this study if they had neurologic or medical conditions that could have potentially affect their central nervous system, such as atypical headaches, head trauma with loss of consciousness, chronic lung disease, kidney disease, chronic hepatic disease, thyroid disease, active cancer, cerebrovascular disease, epilepsy, seizures, substance abuse related disorders, or mental retardation.
SNPs selection, genotyping, and genomic sequencing
2021 Copyright OAT. All rights reserv
This study was designed to examine the association of SCZ patients’ cognitive performance (through WAIS-III implementation [17]) with NTNG genes. Venous blood was collected from the subjects. Genomic DNA was extracted from the whole blood using standard procedures. The SNPs [27-29,31] were genotyped using the TaqMan allelic discrimination assay (Applied Biosystems, Foster City, CA). No deviations from the Hardy-Weinberg equilibrium in the examined SNPs were detected (p > 0.05).
Statistical analysis
All statistical analyses were performed using IBM SPSS Statistics 19.0 software (IBM Japan, Tokyo, Japan). The effects of the diagnosis, genotype and their interaction on cognitive performances in the WAIS were analyzed by two-way analyses of covariance (ANCOVA). Diagnosis and genotype statuses were included in the model as independent variables (ST1). FIQ and each WAIS subscale score (VIQ, PIQ, VC, PO, WM, PS, Vc, SiM, IF, CH, AM, DS, LNS, PC, BD, and MR) were included as dependent variables. Sex and years of education were treated as covariates, as they were possible confounding factors. All p values are two tailed, and statistical significance was defined as *p < 0.05 and **p < 0.01.
Identity percent calculations and the definition of ROIs
The complete procedure is described in the Figures 2-3 legend. Stretcher [71] was used for the alignments (the default values were: gap penalty – 16 (DNA) and 12 (protein), and the extend penalty – 4 (DNA) and 2 (protein)), for the percent identity calculations and evolutionary rates. A ROI was initially defined as a minimal area surrounding a SNP mutation allele incorporating the outmost evolutionary dramatic changes, ±10 nu.
Mouse primates and hominins NTNG paralogs DNA and encoded protein sequences reconstruction. Genomes for mouse (GRC38.p3) and marmoset (C_jacchus3.2.1) were from Ensemble. Since chimpanzee’s genome is based only on a single individual (CHIMP2.1.4, Clint) and contains several questionable information we have reconstructed a consensus genome sequence for both NTNG genes based on 25 primate sequences of Pan troglodytes [72]. All datasets used for the NTNG paralogs DNA and encoded by them proteins reconstruction are listed in ST2. For details refer to SM.
Supplementary materials (SM)
Contain additional Results and Discussion, Supplementary Methods (ancient and primate genomes reconstructions), and Supplementary Tables (ST1 and ST2) as a single compiled pdf file. Also included are human Netrin-G1 and Netrin-G2 alignments, as well as 101 nu alignments for all 11 ROIs across all analysed species.
View supplementary data
Acknowledgements
Authors would like to acknowledge the financial support provided by Funding Program for World-Leading Innovative R&D on Science and Technology (FIRST Programme) and KAKENHI 15H04290 by the Japan Society for the Promotion of Science (JSPS).
References
- Cohen BA, Pilpel Y, Mitra RD, Church GM (2002) Discrimination between paralogs using microarray analysis: application to the Yap1p and Yap2p transcriptional networks. Mol Biol Cell 13: 1608-1614. [Crossref]
- Nakashiba T, Nishimura S, Ikeda T, Itohara S (2002) Complementary expression and neurite outgrowth activity of netrin-G subfamily members. Mech Dev 111: 47-60. [Crossref]
- Nishimura-Akiyoshi S, Niimi K, Nakashiba T, Itohara S (2007) Axonal netrin-Gs transneuronally determine lamina-specific subdendritic segments. Proc Natl Acad Sci U S A 104: 14801-14806. [Crossref]
- Matsukawa H, Akiyoshi-Nishimura S, Zhang Q, Luján R, Yamaguchi K, et al. (2014) Netrin-G/NGL complexes encode functional synaptic diversification. J Neurosci 34: 15779-15792. [Crossref]
- Lai Wing Sun K, Correia JP, Kennedy TE (2011) Netrins: versatile extracellular cues with diverse functions. Development 138: 2153-2169. [Crossref]
- Yu S, Guo Z, Johnson C, Gu G, Wu Q (2013) Recent progress in synthetic and biological studies of GPI anchors and GPI-anchored proteins. Curr Opin Chem Biol 17: 1006-1013. [Crossref]
- Sevcsik E, Brameshuber M, Fölser M, Weghuber J, Honigmann A, et al. (2015) GPI-anchored proteins do not reside in ordered domains in the live cell plasma membrane. Nat Commun 6: 6969. [Crossref]
- Klotzsch E, Schütz GJ (2012) A critical survey of methods to detect plasma membrane rafts. Philos Trans R Soc Lond B Biol Sci 368: 20120033. [Crossref]
- Abitua PB, Wagner E, Navarrete IA, Levine M (2012) Identification of a rudimentary neural crest in a non-vertebrate chordate. Nature 492: 104-107. [Crossref]
- Stolfi A, Ryan K, Meinertzhagen IA, Christiaen L (2015) Migratory neuronal progenitors arise from the neural plate borders in tunicates. Nature 527: 371-374. [Crossref]
- Abitua PB, Gainous TB, Kaczmarczyk AN, Winchell CJ, Hudson C, et al. (2015) The pre-vertebrate origins of neurogenic placodes. Nature 524: 462-465. [Crossref]
- Wise SP (2008) Forward frontal fields: phylogeny and fundamental function. Trends Neurosci 31: 599-608. [Crossref]
- Preuss TM (2012) Human brain evolution: from gene discovery to phenotype discovery. Proc Natl Acad Sci U S A 109: 10709-10716. [Crossref]
- Geschwind DH, Rakic P (2013) Cortical evolution: judge the brain by its cover. Neuron 80: 633-647. [Crossref]
- Izpisua Belmonte JC, Callaway EM, Caddick SJ, Churchland P, Feng G, et al. (2015) Brains, genes, and primates. Neuron 86: 617-631. [Crossref]
- Joshi PK, Esko T, Mattsson H, Eklund N, Gandin I, et al. (2015) Directional dominance on stature and cognition in diverse human populations. Nature 523: 459-462. [Crossref]
- Wechsler D (1958) The Measurement And Appraisal Of Adult Intelligence. (4th edn). The Williams & Wilkins Company. 324.
- Deary IJ, Spinath FM, Bates TC (2006) Genetics of intelligence. Eur J Hum Genet 14: 690-700. [Crossref]
- Gläscher J, Tranel D, Paul LK, Rudrauf D, Rorden C, et al. (2009) Lesion mapping of cognitive abilities linked to intelligence. Neuron 61: 681-691. [Crossref]
- Spearman C (1904) “General intelligence" objectively determined and measured. Am J Psychol 15: 201-292.
- Hampshire A, Highfield RR, Parkin BL, Owen AM (2012) Fractionating human intelligence. Neuron 76: 1225-1237. [Crossref]
- Lett TA, Voineskos AN, Kennedy JL, Levine B, Daskalakis ZJ (2014) Treating working memory deficits in schizophrenia: a review of the neurobiology. Biol Psychiatry 75: 361-370. [Crossref]
- Forbes NF, Carrick LA, McIntosh AM, Lawrie SM (2009) Working memory in schizophrenia: a meta-analysis. Psychol Med 39: 889-905. [Crossref]
- Leeson VC, Robbins TW, Matheson E, Hutton SB, Ron MA, et al. (2009) Discrimination Learning, Reversal, and Set-Shifting in First-Episode Schizophrenia: Stability Over Six Years and Specific Associations with Medication Type and Disorganization Syndrome. Biol Psychiatry 66: 586-593. [Crossref]
- Barnett JH, Robbins TW, Leeson VC, Sahakian BJ, Joyce EM, et al. (2010) Assessing cognitive function in clinical trials of schizophrenia. Neurosci Biobehav Rev 34: 1161-1177. [Crossref]
- Toulopoulou T, Picchioni M, Rijsdijk F, Hua-Hall M, Ettinger U, et al. (2007) Substantial genetic overlap between neurocognition and schizophrenia: genetic modeling in twin samples. Arch Gen Psychiatry 64: 1348-1355. [Crossref]
- Fukasawa M, Aoki M, Yamada K, Iwayama-Shigeno Y, Takao H, et al. (2004) Case-control association study of human netrin G1 gene in Japanese schizophrenia. J Med Dent Sci 51: 121-128. [Crossref]
- Aoki-Suzuki M, Yamada K, Meerabux J, Iwayama-Shigeno Y, Ohba H, et al. (2005) A Family-Based Association Study and Gene Expression Analyses of Netrin-G1 and -G2 Genes in Schizophrenia. Biol Psychiatry 57: 382-393. [Crossref]
- Arinami T, Ohtsuki T, Ishiguro H, Ujike H, Tanaka Y, et al. (2005) Genomewide high-density SNP linkage analysis of 236 Japanese families supports the existence of schizophrenia susceptibility loci on chromosomes 1p, 14q, and 20p. Am J Human Genet 77: 937-944. [Crossref]
- Eastwood SL, Harrison PJ (2008) Decreased mRNA expression of netrin-G1 and netrin-G2 in the temporal lobe in schizophrenia and bipolar disorder. Neuropsychopharmacology 33: 933-945. [Crossref]
- Ohtsuki T, Horiuchi Y, Koga M, Ishiguro H, Inada T, et al. (2008) Association of polymorphisms in the haplotype block spanning the alternatively spliced exons of the NTNG1 gene at 1p13.3 with schizophrenia in Japanese populations. Neurosci Lett 435: 194-197. [Crossref]
- Zakharyan R, Boyajyan A, Arakelyan A, Gevorgyan A, Mrazek F, et al. (2011) Functional variants of the genes involved in neurodevelopment and susceptibility to schizophrenia in an Armenian population. Hum Immunol 72: 746-748. [Crossref]
- Zhu Y, Yang H, Bi Y, Zhang Y, Zhen C, et al. (2011) Positive association between NTNG1 and schizophrenia in Chinese Han population. J Genet 90: 499-502. [Crossref]
- Ayalew M, Le-Niculescu H, Levey DF, Jain N, Changala B, et al. (2012) Convergent functional genomics of schizophrenia: from comprehensive understanding to genetic risk prediction. Mol Psychiatry 17: 887-905. [Crossref]
- Wilcox JA, Quadri S (2014) Replication of NTNG1 association in schizophrenia. Psychiatr Genet 24: 266-268. [Crossref]
- Nakashiba T, Ikeda T, Nishimura S, Tashiro K, Honjo T, et al. (2000) Netrin-G1: a Novel Glycosyl Phosphatidylinositol-Linked Mammalian Netrin That Is Functionally Divergent from Classical Netrins. J Neurosci 20: 6540-6550. [Crossref]
- APA (American Psychiatric Association) (2013) Diagnostic and Statistical Manual of Mental Disorders (DSM-V®). (5th Edn). 991.
- Yoon JH, Sheremata SL, Rokem A, Silver MA (2014) Windows to the soul: vision science as a tool for studying biological mechanisms of information processing deficits in schizophrenia. Front Psychol 4: A681. [Crossref]
- John B, Lewis KR (1966) Chromosome variability and geographic distribution in insects. Science 152: 711-721. [Crossref]
- Gottesman II, McGue M (2015) Endophenotype. In: The Encyclopedia of Clinical Psychology. (1st Edn). Edited by: Cautin RL, Lilienfeld SO. JohnWiley & Sons, Inc.
- Glahn DC, Knowles EE, McKay DR, Sprooten E, Raventós H, et al. (2014) Arguments for the sake of endophenotypes: examining common misconceptions about the use of endophenotypes in psychiatric genetics. Am J Med Genet B Neuropsychiatr Genet 165B: 122-130. [Crossref]
- Frydecka D, Eissa AM, Hewedi DH, Ali M (2014) Impairments of working memory in schizophrenia and bipolar disorder: the effect of history of psychotic symptoms and different aspects of cognitive task demands. Front Behav Neurosci 8: 416. [Crossref]
- Baddeley A (1992) Working memory. Science 255: 556-559. [Crossref]
- Mackintosh NJ (2011) "History of theories and measurement of Intelligence". In: The Cambridge Handbook of Intelligence. Edited by Sternberg RJ, Kaufman SB. 3-19.
- del Rosario RC, Rayan NA, Prabhakar S (2014) Noncoding origins of anthropoid traits and a new null model of transposon functionalization. Genome Res 24: 1469-1484. [Crossref]
- Kellis M, Wold B, Snyder MP, Bernstein BE, Kundaje A, et al. (2014) Defining functional DNA elements in the human genome. Proc Natl Acad Sci U S A 111: 6131-6138. [Crossref]
- Hemani G, Shakhbazov K, Westra HJ, Esko T, Henders AK, et al. (2014) Detection and replication of epistasis influencing transcription in humans. Nature 508: 249-253. [Crossref]
- Plomin R, Deary IJ (2015) Genetics and intelligence differences: five special findings. Mol Psychiatry 20: 98-108. [Crossref]
- Benyamin B, Pourcain B, Davis OS, Davies G, Hansell NK, et al. (2014) Childhood intelligence is heritable, highly polygenic and associated with FNBP1L. Mol Psychiatry 19: 253-258. [Crossref]
- Desrivières S, Lourdusamy A, Tao C (2015) Single nucleotide polymorphism in the neuroplastin locus associates with cortical thickness and intellectual ability in adolescents. Mol Psychiatry 20: 263-274. [Crossref]
- Davies G, Tenesa A, Payton A, Yang J, Harris SE, et al. (2011) Genome-wide association studies establish that human intelligence is highly heritable and polygenic. Mol Psychiatry 16: 996-1005. [Crossref]
- Rietveld CA, Medland SE, Derringer J, Yang J, Esko T, et al. (2013) GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science 340: 1467-1471. [Crossref]
- Prosselkov P, Polygalov D, Zhang Q, McHugh TJ, Itohara S (2015) Cognitive Domains function complementation by NTNG gene paralogs. bioRxiv.
- Prabhakar S, Noonan JP, Pääbo S, Rubin EM (2006) Accelerated evolution of conserved noncoding sequences in humans. Science 314: 786. [Crossref]
- Rubinstein M, de Souza FS (2013) Evolution of transcriptional enhancers and animal diversity. Philos Trans R Soc Lond B Biol Sci 368: 20130017. [Crossref]
- Harmston N, Baresic A, Lenhard B (2013) The mystery of extreme non-coding conservation. Philos Trans R Soc Lond B Biol Sci 368: 20130021. [Crossref]
- Meyer M, Kircher M, Gansauge MT, Li H, Racimo F, et al. (2012) A high-coverage genome sequence from an archaic Denisovan individual. Science 338: 222-226. [Crossref]
- Lazaridis I, Patterson N, Mittnik A, Renaud G, Mallick S, et al. (2014) Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 513: 409-413. [Crossref]
- Keller A, Graefen A, Ball M, Matzas M, Boisguerin V, et al. (2012) New insights into the Tyrolean Iceman's origin and phenotype as inferred by whole-genome sequencing. Nat Commun 3: 698. [Crossref]
- Rasmussen M, Li Y, Lindgreen S, Pedersen JS, Albrechtsen A, et al. (2010) Ancient human genome sequence of an extinct Palaeo-Eskimo. Nature 463: 757-762. [Crossref]
- Appenzeller T (2013) Neanderthal culture: old masters. Nature 497: 302-304. [Crossref]
- Wong K (2015) Neandertal Minds. Scientific American 312: 36-43.
- Gabora L, Russon A (2011) "The evolution of Intelligence". In: “The Cambridge Handbook of Intelligence”. Edited by Sternberg RJ, Kaufman SB. 328-350.
- Balter M (2010) Evolution of behavior. Did working memory spark creative culture? Science 328: 160-163. [Crossref]
- Crabtree GR (2013) Our fragile intellect. Part I. Trends in Genetics 29: 1-3.
- Haak W, Lazaridis I, Patterson N, Rohland N, Mallick S, et al. (2015) Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 522: 207-211. [Crossref]
- Song M, Zhou Y, Li J, Liu Y, Tian L, et al. (2008) Brain spontaneous functional connectivity and intelligence. Neuroimage 41: 1168-1176. [Crossref]
- Pamplona GS, Santos Neto GS, Rosset SR, Rogers BP, Salmon CE (2015) Analyzing the association between functional connectivity of the brain and intellectual performance. Front Hum Neurosci 9: 61. [Crossref]
- Der Sarkissian C, Allentoft ME, Ávila-Arcos MC, Barnett R, Campos PF, et al. (2015) Ancient genomics. Philos Trans R Soc Lond B Biol Sci 370: 20130387. [Crossref]
- Ohi K, Hashimoto R, Nakazawa T, Okada T, Yasuda Y, et al. (2012) The p250GAP gene is associated with risk for schizophrenia and schizotypal personality traits. PLoS One 7: e35696. [Crossref]
- McWilliam H, Li W, Uludag M, Squizzato S, Park YM, et al. (2013) Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res 41: W597-600. [Crossref]
- Prado-Martinez J, Sudmant PH, Kidd JM, Li H, Kelley JL, et al. (2013) Great ape genetic diversity and population history. Nature 499: 471-475. [Crossref]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, et al. (2010) A draft sequence of the Neandertal genome. Science 328: 710-722. [Crossref]
- Prüfer K, Racimo F, Patterson N, Jay F, Sankararaman S, et al. (2014) The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505: 43-49. [Crossref]



