We performed a Cross sectional study to determine the frequency of Philadelphia chromosome and frequency of standard and variant translocation in chronic myeloid leukemia cases by the collaboration of Pathology department of Liaquat University of Medical and Health Sciences, Jamshoro and Isra University Hospital, Hyderabad from May to September 2014. A sample of 145 diagnosed cases of CML was selected according to inclusion. Bone marrow and peripheral Blood samples were collected in sodium heparinized bottles. Cytogenetic analysis was performed by karyotyping according to ISCN guidelines for human cytogenetic nomenclature using cytovision system for image analysis, reverse transcription polymerase chain reaction (RT-PCR) was performed to identify the various BCR- ABL transcripts. Ph+ and Ph- chromosomes were noted in 133 (91.7) and 12 (8.2%) of cases respectively. Of 133 Ph+ chromosome, standard chromosome was noted in 121 (90.9%), simple variant in 9 (6.7) and complex variants were noted in 3 (2.2%) of cases. Ph + 133 (91.7%) showed bcr-abl positivity in all subjects. Of 12 (8.2%) Ph- subjects, 7 (%) were bcr-abl positive and 5 (%) were bcr-able negative. Sensitivity and specificity of bcr-abl transcripts was calculated at 95% and 100% respectively. The present study reports Philadelphia chromosome in 90.9% and variant cytogenetic abnormalities are low similar to reported from other countries. Proper assessment of the variant translocations requires better categorization of these translocations.
philadelphia chromosome, chronic myeloid leukemia, sindh
Chronic myeloid leukemia (CML) is a myeloproliferative disorder characterized by the presence of the Philadelphia chromosome (Ph) which is the derivative chromosome 22 of the translocation t (9; 22) (q34.1; q11.2). Due to this rearrangement, the break-point cluster region (BCR) gene at position 22q11 [1,2] is juxtaposed to the c-Abelson (ABL1) gene at 9q34, resulting in the BCR-ABL1 fusion gene, encoding a constitutively active tyrosine kinase protein. The identification of this abnormality is important for the diagnosis of the disease as determined by the WHO Tumor Classification1and for treatment purposes. The first therapeutic choice, tyrosine kinase inhibitors, has shown great therapeutic efficacy [2]. The Ph is detected by G-band karyotyping in around 90% of CML patients among whom 5–10% may have variant types [3-5]. Variant Ph chromosomes are characterized by the involvement of another chromosome in addition to chromosome 9 or 22. It can be a simple type of variant when only one additional chromosome is involved, or complex, in which two or more chromosomes, besideschromosomes 9 and 22, take part in the translocation [6,7]. Variant Ph breakpoints occur in hotspots across the genome, usually in the G-light bands, within the cytosine and guanine (CG) richest parts of the genome [8]. However, the mechanism of variant Ph generation and the molecular bases of biological differences between classic Ph and variant Ph chromosomes are not fully understood [9]. Recently, Albano et al. [10] reported a study they performed on gene expression profiling (GEP) using microarrays to identify some of these differences [10].
The prognostic significance of variant Ph chromosomes has already been discussed [11] and it has been shown that the variant aberration does not impact on cytogenetic or molecular responses or even on clinical outcome. Variant Ph chromosomes are distinguished from additional chromosomal abnormalities or clonal evolution that drives disease progression. The clonal evolution is a reflection of a genetic instability that characterizes the transition to advanced phase [12]. In this situation, i (17q), a second Ph and +8 are frequently found [1]. A search of local literature showed only a few studies had been conducted and reported, hence there is an urgent need to conduct more studies to explore cytogenetic and molecular abnormalities in chronic myeloid leukemia cases in our local population of Sind.
This cross sectional study was conducted at pathology department of Liaquat University of Medical and Health Sciences, Jamshoro and Isra University Hospital, Hyderabad from May to September 2014. A sample of 145 newly diagnosed cases of CML was included. Patients with acute leukemia, polycythemia, essential thrombocythemia and myelofibrosis, multiple myeloma were excluded from the study. Diagnosed cases of CML were included, while patients with acute leukemia, lymphoma, and multiple myeloma. Written informed consent was taken from the patients. The study was approved by the ethical committee of Isra University Hyderabad.
Bone marrow and Blood samples were collected in sodium heparinized vacutainer by applying appropriate techniques. CBC was performed on automatic hematoanalyzer, Sysmex XN 1000i. Cytogenetic analysis was performed by karyotyping according to ISCN guidelines for human cytogenetic nomenclature using cytovision system for image analysis, reverse transcription polymerase chain reaction (RT-PCR) was performed to identify the various BCR- ABL transcripts by Qi guine kits.
The data was analyzed on SPSS version 21.0 (IBM, Corporation) and Microsoft excel. The continuous variables were presented as mean ± SD and analyzed using student’s t-test. Categorical variables were analyzed by Chi-square test and results were presented as frequencies and percentages. Data was presented in tables, graphs and charts. P-value of ≤ 0.05 was defined significant.
Mean ± SD of age was noted as 36 ± 11.7 years. Of 145 cases, most frequent age groups were 20-29.9 and 30-39.9 noted in 47.5% and 44.8% respectively. Of 145 cases, 109 (75.1%) were male and 36 (24.8) were female (p=0.001). Anemia was noted in 74.4% and hematocrit (<20%) in 46.2% of cases. Leukocytosis of >50,000/µL was noted in 68.5% of total cases.
Ph+ and Ph- chromosome was noted in 133 (91.7) and 12 (8.2%) of cases respectively (Figure 1). Of 133 Ph+ chromosome, standard chromosome was noted in 121 (90.9%), simple variant in 9 (6.7%) and complex variants were noted in 3 (2.2%) of cases, shown in Table 1 and Figure 2. Simple and complex variant translocations are shown in table 1. Ph + 133 (91.7) showed bcr-abl positivity in all subjects. Of 12 (8.2%) Ph- subjects, 7 (%) were bcr-abl positive and 5 (%) were Bcr-Abl negative (Figure 3). Sensitivity and specificity of bcr-Abel transcripts were calculated at 95% and 100% respectively.
Table 1. Simple and complex variants of Philadelphia chromosome (n=12)
|
Variant Ph chromosome |
Chromosome translocation |
1. |
Simple variant Ph |
46 xy t(16;22) |
2. |
Simple variant Ph |
46 xx t(19;22) |
3. |
Simple variant Ph |
46 xx t(13;22) |
4. |
Simple variant Ph |
46 xy t(17;22) |
5. |
Simple variant Ph |
46 xx t(11;22) |
6. |
Simple variant Ph |
46 xx t(18;22) |
7. |
Simple variant Ph |
46 xy t(15;22) |
8. |
Simple variant Ph |
46 xx t(14;22) |
9. |
Simple variant Ph |
46 xx t(12;22) |
10. |
Complex variant Ph |
46 xy t(6;9;22) |
11. |
Complex variant Ph |
46 xx t(5;9;22) |
12. |
Complex variant Ph |
46 xx t(7;9;22) |
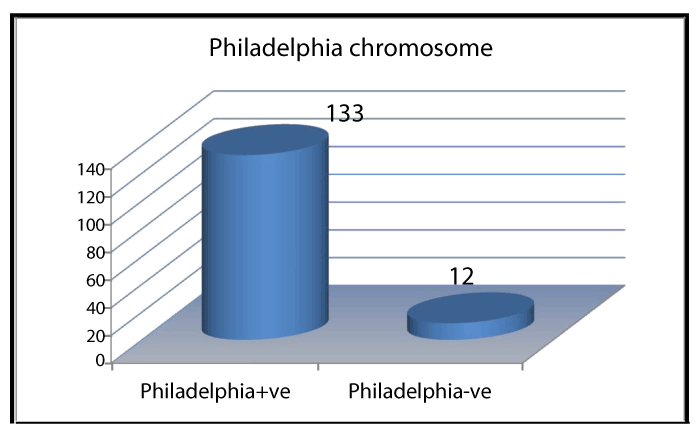
Figure 1. Frequency of Philadelphia chromosome
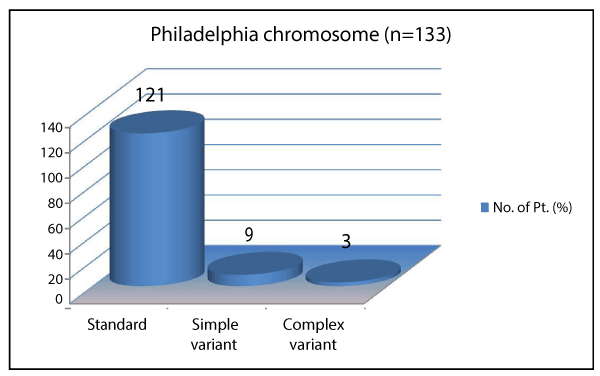
Figure 2. Frequency of Philadelphia chromosome variants
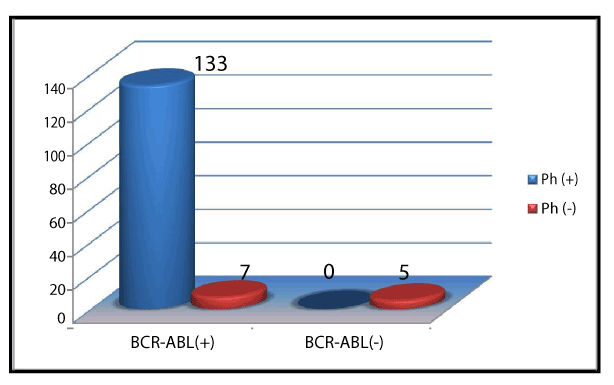
Figure 3. Frequency of bcr-abl transcripts in study population
Cytogenetic and molecular analysis of transcripts of CML – chronic, accelerated and blast phases are shown in Table 2. Most frequent cytogenetic abnormalities were B3a2, b2a2 and b3a2+b2a2 found in 70%, 34% and 2% of cases respectively.
Table 2. Cytogenetic and molecular analysis of transcripts in cases
Transcript |
Chronic Phase |
Accelerated |
Blast |
|
(n=130) |
Phase (n=9) |
Crisis(n=6) |
b3a2 |
70% |
3% |
- |
b2a2 |
24% |
- |
1% |
b3a2+ b2a2 |
34% |
- |
- |
b3a2+e 19a2 |
2% |
- |
- |
b2a2+e 19a2 |
- |
- |
2% |
CML is a prototype myeloproliferative disorder of bone marrow. CML is one of leukemia which may be diagnosed by typical clinical, hematological and morphological features when interpreted in a proper clinical context. Philadelphia chromosomes (Ph) is a sine qua non of CML. Philadelphia chromosome and bcr-abl cytogenetic abnormalities must be detected in all cases of chronic, accelerated and blast crisis phases of CML [13-15].
In the present study, pH chromosome was detected in 90.9% of cases, the finding consistent with previous studies which had reported Ph chromosome in 90-95% of cases [14-16]. Additional cytogenetic abnormalities were detected in 70% of cases in present study as shown in Table 2.
The findings are in full agreement with previous studies which had reported a frequency of 70- 80% [15,16]. Most frequent cytogenetic abnormalities were b3a2, b2a2 and b3a2+b2a2 found in 70%, 34% and 2% of cases respectively (Table 2). The findings are in keeping with previous studies from Western countries [17-21]. However, another previous study had reported less frequency of b3a2 and b2a2 in 55% and 40% [6] of cases [22] respectively. The differences might have been introduced due to sample size, sampling techniques, study designs and nonetheless geographical differences. Paz-y-Miño et al. found frequencies of 5.4% for the b3a2 transcripts and 94.6%for the b2a2 transcripts in Ecuadorian Mestizos CML patients. Findings are in contrast to present study (Table 2). Rosas-Cabral et al. detected b3a2 BCR-ABL transcripts in 28% cases, b2a2 in 59% cases, and 13% with both b3a2/b2a2 transcripts among 97 Philadelphia-positive CML Mexican cases. The findings are inconsistent with present study (Table 2). Ruiz-Argüelles et al. studied a group of 238 Mexican Mestizos patients with Ph positive CML; 54.2 per cent showed b3a2 subtype, 43.2% b2a2 and 2.5% b3a2/b2a2. The findings are consistent with present study (Table 2). De Lemos et al. from Brazil performed RT-PCR for BCR-ABL in 22 CML patients. Of these patients, 15 (68%) were in chronic phase, five (23%) in accelerated phase and two (9%) in blastic phase; 59% patients had b3a2 and 41% had b2a2 transcripts. The findings of this previous study are very close to the findings of present study. Yaghmaieet al. studied 75 adult Iranian CML patients; 83% patients expressed one of the p210 BCR-ABL transcripts (b3a2, 63% and b2a2, 20%), while the remaining showed one of the transcripts of b3a3, b2a3, e1a2 or co-expression of b3a2 and b2a2. b3a2 and b2a2 were co-expressed in 5% patients. Mondal et al. [23] studied 122 CML patients and details of cytogenetic analysis are not provided. Out of 122, 112 (91.8%) patients were positive for one or more of the four junctional types tested. Findings of above studies are consistent with present study. Polampalli et al. [24] studied 202 CML patients; 138 (68%) had the b3a2 type bcr-able transcript which is consistent to 70% in present study. 64 (32%) had the b2a2 type which is also in keeping with our finding of 34% as shown in Table 2. Ph positivity in present study is in comparison to previous studies reported from India [25-28]. Present study reports Philadelphia chromosome, its variants and cytogenetic abnormalities which are consistent with most of studies mentioned in literature. Strength of present study lies in its prospective design, inclusion and exclusion criteria. Limitations of present may be small sample size and we could not analyze all of cytogenetic abnormalities due to lack of modern laboratory facilities and funding issues.
The present study reports Philadelphia frequency of 90.9% and cytogenetic abnormalities similar to reported from other countries. Cytogenetic and molecular studies must be conducted for better management of CML cases in our locality. Overall, this study provides further understanding of cytogenetic and molecular abnormalities in CML cases.
All authors declare that they do not have any conflict of interests to declare.
We acknowledge the clinical staff at Israuniversity and Liaquat university who painstakingly helped in this project. We are thankful to Dr. Khaskali who helped us shape this manuscript.
- Chauffaille Mde L, Bandeira AC, da Silva AS (2015) Diversity of breakpoints of variant Philadelphia chromosomes in chronic myeloid leukemia in Brazilian patients. Rev Bras Hematol Hemoter 37: 17-20. [Crossref]
- Baccarani M, Deininger MW, Rosti G, Hochhaus A, Soverini S, et al. (2013) European LeukemiaNet recommendations for the management of chronic myeloid leukemia: 2013. Blood 122: 872-884. [Crossref]
- Ishihara T, Minamihisamatsu M (1988) The Philadelphia chromosome. Considerations based on studies of variant Ph translocations. Cancer Genet Cytogenet 32: 75-92. [Crossref]
- Fisher AM, Strike P, Scott C, Moorman AV (2005) Breakpoints of variant 9;22 translocations in chronic myeloid leukemia locate preferentially in the CG-richest of the genome. Genes Chromosomes Cancer 43: 383-389. [Crossref]
- Marzocchi G, Castagnetti F, Luatti S, Baldazzi C, Stacchini M, et al. (2011) Variant Philadelphia translocations: molecular-cytogenetic characterization and prognostic influence on frontline imatinib therapy, a GIMEMA Working Party on CML analysis. Blood 117: 6793-6800. [Crossref]
- Sandberg AA (1980) Chromosomes and causation of human cancer and leukemia. XL. The Ph1 and other translocations in CML. Cancer 46: 2221-2226. [Crossref]
- Heim S, Mittelman F (1995) Cancer cytogenetics. (2nd edn). New York: Wiley Liss.
- Jeffs AR, Benjes SM, Smith TL, Sowerby SJ, Morris CM (1998) The BCR gene recombines preferentially with Alu elements in complex BCR-ABL translocations of chronic myeloid leukaemia. Hum Mol Genet 7: 767-776. [Crossref]
- Chauffaille ML (2008) Cytogenetics and FISH monitoring CMLduring tyrosine kinase inhibitors treatment. Rev Bras Heamtol Hemoter 30: 13-19. [Crossref]
- Albano F, Zagaria A, Anelli L, Coccaro N, Impera L, et al. (2013) Gene expression profiling of chronic myeloid leukemia with variant t(9;22) reveals a different signature from cases with classic translocation. Mol Cancer 12: 36. [Crossref]
- Sweet K, Zhang L, Pinilla-Ibarz J (2013) Biomarkers for determining the prognosis in chronic myelogenous leukemia. J Hematol Oncol 6: 54. [Crossref]
- Quintás-Card2021 Copyright OAT. All rights reservogy of bcr-abl1-positive chronic myeloid leukemia. Blood 113: 1619-1630. [Crossref]
- Anand MS, Varma N, Varma S, Rana KS, Malhotra P (2012) Cytogenetic & molecular analyses in adult chronic myelogenous leukaemia patients in north India. Indian J Med Res 135: 42-48. [Crossref]
- Vardiman JW, Melo JV, Baccarani M, Thiele J (2008) Myeloproliferative Neoplasms. In: Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H. WHO classification of tumours of haematopoietic and lymphoid tissues. Lyon: IARC Press: 32-37.
- Rabinowitz I, Larson RS (2004) Chronic myeloid leukemia. In: Greer JP, Foerster J, Lukens JN, Rodgers GM, Paraskevas F, et al.. Wintrobe's clinical hematology. (11th edn). Philadelphia: Lippincott Williams & Wilkins: 2235-2258.
- Simons CM, Stratton CW, Kim AS (2011) Peripheral blood eosinophilia as a clue to the diagnosis of an occult Coccidioides infection. Hum Pathol 42: 449-453. [Crossref]
- Paz-y-Miño C, Burgos R, Morillo SA, Santos JC, Fiallo BF, et al. (2002) BCR-ABL rearrangement frequencies in chronic myeloid leukemia and acute lymphoblastic leukemia in Ecuador, South America. Cancer Genet Cytogenet 132: 65-67. [Crossref]
- Rosas-Cabral A, Martínez-Mancilla M, Ayala-Sánchez M, Vela-Ojeda J, Bahena-Reséndiz P, et al. (2003) Analysis of Bcr-abl type transcript and its relationship with platelet count in Mexican patients with chronic myeloid leukemia. Gac Med Mex 139: 553-559. [Crossref]
- Ruiz-Argüelles GJ, Garcés-Eisele J, Reyes-Núñez V, Ruiz-Delgado GJ (2004) Frequencies of the breakpoint cluster region types of the BCR/ABL fusion gene in Mexican Mestizo patients with chronic myelogenous leukemia. Rev Invest Clin 56: 605-608. [Crossref]
- de Lemos JA, de Oliveira CM, Scerni AC, Bentes AQ, Beltrão AC, et al. (2005) Differential molecular response of the transcripts B2A2 and B3A2 to imatinib mesylate in chronic myeloid leukemia. Genet Mol Res 4: 803-811. [Crossref]
- Yaghmaie M, Ghaffari SH, Ghavamzadeh A, Alimoghaddam K, Jahani M, et al. (2008) Frequency of BCR-ABL fusion transcripts in Iranian patients with chronic myeloid leukemia. Arch Iran Med 11: 247-251. [Crossref]
- Melo JV (1996) The diversity of BCR-ABL fusion proteins and their relationship to leukemia phenotype. Blood 88: 2375-2384. [Crossref]
- Mondal BC, Bandyopadhyay A, Majumdar S, Mukhopadhyay A, Chandra S, et al. (2006) Molecular profiling of chronic myeloid leukemia in eastern India. Am J Hematol 81: 845-849. [Crossref]
- Polampalli S, Choughule A, Negi N, Shinde S, Baisane C, et al. (2008) Analysis and comparison of clinicohematological parameters and molecular and cytogenetic response of two Bcr/Abl fusion transcripts. Genet Mol Res 7: 1138-1149. [Crossref]
- Jacob RT, Gayathri K, Surath A, Rao DR (2002) Cytogenetic profile of chronic myeloid leukemias. Indian J Cancer 39: 61-65. [Crossref]
- VinSheth FJ, Sheth JJ, Patel AI, Shah AD, Verhest A (2002) Usefulness of cytogenetics in leukemias. Indian J Cancer 39: 139-142. [Crossref]
- Chavan D, Ahmad F, Iyer P, Dalvi R, Kulkarni A, et al. (2006) Cytogenetic investigation in chronic myeloid leukemia: study from an Indian population. Asian Pac J Cancer Prev 7: 423-426. [Crossref]
- Bakshi SR, Brahmbhatt MM, Trivedi PJ, Shukla SN, Shah PM (2006) Atypical D-FISH patterns of BCR/ABL gene rearrangements in 169 chronic myeloid leukemia patients. J Assoc Genet Technol 32: 164-167. [Crossref]



