Mycoplasma bovis, a serious cause of bovine respiratory disease often associated with other bacteria and viruses, is prevalent worldwide including Turkey. It is difficult to control because antibiotics are becoming increasingly ineffective and commercial vaccines are not available. The aims of this study were to detect infective agents causing respiratory disease in a calf rearing unit in Turkey to determine antibiotic susceptibilities of the strains isolated and to carry out molecular typing. In diseased lungs from calves dying of pneumonia we found M. bovis to be present in all samples of acute and chronic diseased tissue. Mannheimia haemolytica was detected mostly in chronic tissue (39 %) while the reverse was true for Pasteurella multocida with most isolations from acute tissue (25 %). High minimum inhibitory concentrations (MIC) were seen for most antibiotics against M. bovis: tylosin, tilmicosin, erythromycin, chloramphenicol, oxytetracycline and ciprofloxacin with MIC50s of >32 μg/ml, intermediate-sensitivity to florfenicol, spectinomycin, danofloxacin, enrofloxacin and marbofloxacin with MIC50 of 8 μg/ml, and susceptible to lincomycin, clindamycin and tulathromycin with MIC50 of 1, 0.25, 0.25 μg/ml respectively. While all tested isolates of both M. haemolytica and P. multocida were resistant to gentamicin, P. multocida isolates showed resistance rates of 100 % to erythromycin and tylosin, 88 % to trimethoprim-sulphametoxasol, 75% to tetracycline and tilmicosin, 50 % to tulathromycin and enrofloxacin. The resistance rates of M. haemolytica isolates were 90 % to erythromycin, 75 % to tylosin, 64 % to tetracycline, 55 % to trimethoprim-sulphametoxasol, 36 % to tilmicosin, 18 % to enrofloxacin, 9 % to marbofloxacin, florfenicol, ampicillin and penicillin. Finally, M. bovis could be divided into two distinct genetic clusters by molecular typing tests.
Mycoplasma bovis, Mannheimia haemolytica, Pasteurella multocida, calf, pneumonia, antibiotic susceptibility
Diseases of the bovine respiratory system are causes of major economical losses in cattle farms worldwide. Bovine pneumonic pasteurellosis, also known as bovine respiratory disease (BRD), bovine enzootic bronchopneumonia or respiratory disease complex of cattle, is a multifactorial disorder caused by a combination of one or more viruses, bacteria and mycoplasmas. It is estimated that BRD is one of the most important diseases in the cattle industry with a global mortality and morbidity that are estimated to exceed 1% and 10% of young cattle respectively [1].
Mycoplasma bovis is the most important and most pathogenic bovine mycoplasma worldwide. It was reported that M. bovis is responsible for 25-33 % of all calf pneumonias in Europe [2] and the most serious pathogen in the feedlots of North America [3]. In 2006 the first reports from western Turkey described the isolation of M. bovis from just under a third of lungs of calves affected by pneumonia in a calf rearing unit [4]. Later studies confirmed the presence of M. bovis throughout Turkey [5].
M. bovis is frequently associated with other pathogenic microorganisms such as bovine respiratory syncytial virus (BRSV), parainfluenza 3 virus (PI-3), bovine adenoviruses, bovine viral diarrhoea virus (BVDV), bovine herpesvirus 1 (BoHV1), Pasteurella multocida, Mannheimia haemolytica, Arcanobacterium pyogenes, Histophilus somni, Mycoplasma dispar, Mycoplasma canis and Ureaplasma diversum. These infective agents usually exacerbate disease initiated by M. bovis [2].
Control of bovine mycoplasmosis is hampered by the increasing resistance of M. bovis to antibiotics like the tetracyclines and macrolides routinely used for respiratory disease [6,7] Furthermore no commercial vaccines are presently available. An inactivated vaccine for calf pneumonia caused by M. bovis reduced clinical disease, weight loss, lung lesions and spread to internal organs under experimental conditions [8]. Some success was seen in the UK with an autogenous vaccine which reduced mortality and treatment costs on some calf units [9]. However M. bovis has been shown to be genetically diverse by molecular typing tests [10,11] so it is not certain that a vaccine will protect against all strains.
The aims of this study were to detect infective agents causing respiratory disease in a calf rearing unit in Turkey in 2015, to determine antibiotic susceptibilities of the strains isolated and to carry out molecular typing of M. bovis strains wıth the ultimate aim of producing an inactivated autogenous vaccine.
All diagnostic work was carried out at the Veterinary Control and Research Institute Pendik in between March and September in 2015.
Samples
Lungs of 20 calves with acute pneumonia and lungs of 23 calves with chronic pneumonia based on histopathological findings were subjected to bacterial examination. All samples belonged to calves in a calf rearing unit, originating from multiple farms in Turkey.
Isolation and identification
The lung samples were inoculated onto chocolate agar for the isolation of H. somni [12] and onto 5 % sheep blood agar plates (Oxoid) for the isolation of P. multocida and M. haemolytica [1]. Suspect colonies were further purified and identified using an automated Vitek 2 System.
Mycoplasma isolation and identification was attempted from post mortem samples. Samples for culture included material from lung areas at the interface between healthy tissue and lesions [13]. Isolates were grown in liquid and solid Eaton’s mycoplasma media. Semi-solid agar plates were inoculated with a loop full of the broth from cultures showing mycoplasma growth. M. bovis was identified by growth inhibition test using specific rabbit antiserum [14] and confırmed by PCR/denaturing gradient gel electropheresis (DGGE) [10].
Viral antigen detection
The Bio-X Pulmotest tetra ELISA kit was used for PI, BRSV, BoHV1 and BVDV antigens in lung tissue. Detection of adenovirus 3 antigen was performed using Bio-X Adenovirus 3 ELISA kit.
Typing by pulse field gel electrophoresis (PFGE)
The PFGE was performed for genotyping of M. bovis isolates as described previously [15]. Briefly 10 ml of stationary-phase cultures were harvested by centrifugation, washed and resuspended in buffer. Agarose plugs were incubated in lysis buffer then washed. Slices were cut aseptically from the plugs and equilibrated in restriction buffer for 1 h. Restriction digestion was performed with SmaI according to the manufacturer’s instructions. The fragments were resolved on 1% pulsed-field-agarose gels with a CHEF-DRIII system at 6 V/cm. Gels were stained with ethidium bromide and photographed under UV light. A lambda ladder PFGE marker was used for fragment size determination.
Antimicrobial susceptibility testing for conventional bacteria
Susceptibility testing by the disk diffusion method was performed by means of the direct inoculation method on Mueller Hinton II agar [16]. The antibiotics used are listed in table 1.
Minimum inhibitory concentration test (MIC) for M. bovis
The MIC test was performed by microdilution method [6]. The antibiotics used are listed in table 2. While no offıcial criterion for antimicrobial susceptibilty testing exists for animal mycoplasmas as yet we used the guidelines of Ayling et al., [6] for human mycoplasmas. Strains with MIC values of <2 were considered susceptible; >2 to 8 intermediate susceptible and >8 as resistant.
Bacterial detection
Twenty acute pneumonic lung samples and 23 chronic pneumonic lung samples from calves in a calf rearing unit were examined for the presence of M. bovis and other respiratory disease pathogens. While M. bovis was isolated from all samples, the isolation rate of M. haemolytica was 10 % (2 cases) and P. multocida was 25 % (5 cases) in the acute pneumonic samples. In the test for BVDV antigen, 18 % (2 cases) of the acute pneumonic samples were found positive. The isolation rates in the chronic pneumonic samples were 39 % (9 cases) for M. haemolytica and 13 % (3 cases) for P. multocida. BVDV antigen was detected in 18 % (2 cases) of chronic pneumonic samples. H. somni was not isolated from any of the samples.
Antimicrobial susceptibility testing
While all isolates of both M. haemolytica and P. multocida were resistant to gentamicin, P. multocida isolates showed resistance rates of 100 % to erythromycin and tylosin, 88 % to trimethoprim-sulphametoxasol, 75% to tetracycline and tilmicosin, and 50 % to tulathromycin and enrofloxacin. The resistance rates of M. haemolytica isolates were 90 % to erythromycin, 73 % to tylosin, 64 % to tetracycline, 55 % to trimethoprim-sulphametoxasol, 36 % to tilmicosin, 18 % to enrofloxacin, 9 % to marbofloxacin, florfenicol, ampicillin and penicillin. Higher antibiotic resistance was observed in the isolates from acute cases than in those from chronic cases (Table 1).
Table 1. Antimicrobial susceptibility testing of M. haemolytica and P. multocida strains
No |
Strains |
AMP |
AMX |
PEN |
GEN |
TET |
ENR |
ERY |
SXT |
TUL |
TIL |
TYL |
MBF |
FLO |
CTF |
R |
I |
S |
R |
I |
S |
R |
I |
S |
R |
I |
S |
R |
I |
S |
R |
I |
S |
R |
I |
S |
R |
I |
S |
R |
I |
S |
R |
I |
S |
R |
I |
S |
R |
I |
S |
R |
I |
S |
R |
I |
S |
1 |
A*13/08 M. haemolytica |
|
|
x |
|
|
x |
x |
|
|
* |
|
|
x |
|
|
|
x |
|
x |
|
|
x |
|
|
x |
|
|
x |
|
|
x |
|
|
x |
|
|
x |
|
|
* |
* |
* |
2 |
A14/08 P. multocida |
|
|
x |
|
|
x |
|
|
x |
x |
|
|
x |
|
|
|
x |
|
x |
|
|
x |
|
|
x |
|
|
x |
|
|
x |
|
|
* |
* |
* |
|
|
x |
* |
* |
* |
3 |
A15/08 P. multocida |
|
|
x |
|
|
x |
|
|
x |
x |
|
|
x |
|
|
|
x |
|
x |
|
|
x |
|
|
x |
|
|
x |
|
|
x |
|
|
* |
* |
* |
|
|
x |
* |
* |
* |
4 |
A16/08 P. multocida |
|
x |
|
|
x |
|
|
|
x |
x |
|
|
x |
|
|
x |
|
|
x |
|
|
x |
|
|
x |
|
|
x |
|
|
x |
|
|
* |
* |
* |
|
x |
|
|
|
x |
5 |
A17/08 P. multocida |
|
|
x |
|
x |
|
|
|
x |
x |
|
|
x |
|
|
x |
|
|
x |
|
|
x |
|
|
x |
|
|
x |
|
|
x |
|
|
* |
* |
* |
|
|
x |
|
|
x |
6 |
A18/08 M. haemolitica |
|
x |
|
|
|
x |
|
|
x |
x |
|
|
x |
|
|
|
x |
|
x |
|
|
|
x |
|
|
|
x |
|
|
x |
x |
|
|
|
|
x |
|
|
x |
|
|
x |
A18/08 P. multocida |
|
x |
|
|
|
x |
|
|
x |
x |
|
|
x |
|
|
|
x |
|
x |
|
|
|
x |
|
|
|
x |
x |
|
|
x |
|
|
|
|
x |
|
|
x |
|
|
x |
7 |
C**1/08 M. haemolytica |
|
|
x |
|
|
x |
|
|
x |
x |
|
|
x |
|
|
x |
|
|
* |
* |
* |
x |
|
|
|
|
x |
|
|
x |
x |
|
|
|
|
x |
|
|
x |
|
|
x |
8 |
C2/08 M. haemolytica |
|
|
x |
|
|
x |
|
|
x |
x |
|
|
x |
|
|
|
|
x |
* |
* |
* |
x |
|
|
|
|
x |
x |
|
|
x |
|
|
|
|
x |
|
|
x |
|
|
x |
9 |
C8/08 M. haemolytica |
|
x |
|
|
x |
|
|
x |
|
x |
|
|
x |
|
|
|
x |
|
* |
* |
* |
x |
|
|
|
|
x |
|
|
x |
x |
|
|
|
|
x |
|
|
x |
|
|
x |
10 |
C11/08 M. haemolytica |
x |
|
|
|
|
x |
|
x |
|
x |
|
|
x |
|
|
x |
|
|
* |
* |
* |
x |
|
|
|
|
x |
|
|
x |
x |
|
|
|
|
x |
|
|
x |
|
|
x |
11 |
C13/08 P. multocida |
|
|
x |
|
|
x |
|
|
x |
x |
|
|
|
x |
|
x |
|
|
x |
|
|
x |
|
|
|
|
x |
|
|
x |
x |
|
|
|
|
x |
* |
* |
* |
|
|
x |
12 |
C14/08 M. haemolytica |
|
x |
|
|
x |
|
|
x |
|
x |
|
|
|
x |
|
|
x |
|
|
x |
|
x |
|
|
|
|
x |
x |
|
|
x |
|
|
* |
* |
* |
|
|
x |
|
x |
|
13 |
C17/08 M. haemolytica |
|
|
x |
|
|
x |
|
|
x |
x |
|
|
|
|
x |
|
x |
|
* |
* |
* |
|
x |
|
|
|
x |
|
|
x |
|
|
x |
* |
* |
* |
|
|
x |
|
|
x |
14 |
C18/08 M. haemolitica |
|
|
x |
|
|
x |
|
|
x |
x |
|
|
|
|
x |
|
x |
|
* |
* |
* |
|
x |
|
|
|
x |
|
|
x |
x |
|
|
|
|
x |
|
|
x |
|
|
x |
15 |
C19/08 P. multocida |
|
x |
|
|
x |
|
|
x |
|
x |
|
|
x |
|
|
|
x |
|
x |
|
|
x |
|
|
|
|
x |
x |
|
|
x |
|
|
* |
* |
* |
|
|
x |
|
|
x |
16 |
C21/08 M. haemolitica |
|
|
x |
|
|
x |
|
|
x |
x |
|
|
x |
|
|
|
x |
|
x |
|
|
x |
|
|
x |
|
|
|
|
x |
|
|
x |
* |
* |
* |
|
|
x |
|
|
x |
C21/08 P. multocida |
|
|
x |
|
|
x |
|
|
x |
x |
|
|
|
x |
|
x |
|
|
x |
|
|
x |
|
|
|
|
x |
|
|
x |
x |
|
|
|
|
x |
* |
* |
* |
|
|
x |
17 |
C22/08 M. haemolytica |
|
|
x |
|
|
x |
|
|
x |
x |
|
|
|
|
x |
|
x |
|
* |
* |
* |
|
x |
|
|
|
x |
x |
|
|
|
|
x |
|
|
x |
|
|
x |
|
|
x |
A* : acute pneumonic lung
C**: chronic pneumonic lung
R: resistant
I: intermediate
S: sensitive
* not tested
AMP: Ampicillin, AMX: Amoxycillin, CTF: Ceftifiour, ENR: Enrofloxacin, ERY: Erythromycin, FLO: Florfenicol, GEN: Gentamycin, MBF: Marbofloxacine, PEN: Penicillin, SXT: Trimethoprim- Sulphametoxasol, TET: Tetracycline, TIL: Tilmicosin, TUL: Tulathromycin, TYL: Tylosin
MIC for M. bovis
The isolates were found to be resistant to tylosin, tilmicosin, erythromycin, chloramphenicol oxytetracycline and ciprofloxacin with MIC50s of >32 μg/ml, intermediate-susceptible to florfenicol, spectinomycin, danofloxacin, enrofloxacin and marbofloxacin with MIC50 of 8 μg/ml, and susceptible to lincomycin, clindamycin and tulathromycin with MIC50 of 1, 0.25, 0.25 μg/ml respectively (Table 2).
Table 2. Minimum inhibitory concentrations of M. bovis strains (µg/ml)
M. bovis strains |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
14 |
TYL |
TIL |
LCM |
CLI |
ERY |
CHL |
FLO |
SPT |
OTC |
DFX |
ENR |
CIP |
TUL |
MBF |
BC01/08 |
32 |
>32 |
2 |
0.5 |
>32 |
32 |
8 |
8 |
32 |
8 |
8 |
32 |
1 |
8 |
BC02/08 |
32 |
>32 |
1 |
0.25 |
>32 |
32 |
8 |
8 |
32 |
8 |
32 |
32 |
0.5 |
>8 |
BA 03/08 |
8 |
>32 |
0.12 |
0.12 |
32 |
8 |
8 |
2 |
8 |
0.5 |
2 |
2 |
0.25 |
0.25 |
BC04/08 |
8 |
>32 |
0.25 |
0.12 |
32 |
8 |
8 |
8 |
32 |
8 |
8 |
32 |
0.25 |
8 |
BA05/08 |
8 |
>32 |
0.25 |
0,12 |
8 |
8 |
8 |
2 |
32 |
8 |
8 |
32 |
0.25 |
8 |
BC06/08 |
1 |
>32 |
1 |
8 |
>32 |
8 |
8 |
8 |
32 |
8 |
8 |
8 |
1 |
8 |
BA07/08 |
8 |
>32 |
0.12 |
0.12 |
>32 |
8 |
8 |
8 |
8 |
8 |
8 |
32 |
0.5 |
2 |
BA08/08 |
8 |
>32 |
0.25 |
0.25 |
0.12 |
8 |
8 |
2 |
32 |
8 |
8 |
32 |
0.25 |
0.25 |
BC09/08 |
8 |
>32 |
0.12 |
0.12 |
8 |
8 |
2 |
2 |
8 |
2 |
8 |
32 |
0.25 |
1 |
BA10/08 |
8 |
>32 |
0.5 |
0.12 |
32 |
8 |
8 |
8 |
32 |
8 |
8 |
32 |
0.25 |
8 |
BC11/08 |
32 |
>32 |
1 |
0.25 |
>32 |
32 |
8 |
8 |
>32 |
8 |
8 |
32 |
0.25 |
8 |
BA12/08 |
32 |
>32 |
1 |
0.25 |
>32 |
32 |
8 |
8 |
>32 |
8 |
8 |
32 |
0.25 |
8 |
BA14/08 |
32 |
>32 |
2 |
0.5 |
>32 |
32 |
8 |
8 |
>32 |
8 |
8 |
8 |
0.5 |
2 |
BA15/08 |
32 |
>32 |
1 |
0.25 |
>32 |
32 |
8 |
8 |
>32 |
8 |
8 |
32 |
0.25 |
8 |
BC16/08 |
32 |
>32 |
1 |
0.25 |
>32 |
32 |
8 |
8 |
32 |
1 |
1 |
2 |
0.25 |
1 |
BA17/08 |
8 |
>32 |
1 |
0.12 |
>32 |
32 |
8 |
8 |
>32 |
8 |
8 |
32 |
0.25 |
8 |
BA19/08 |
8 |
>32 |
0.5 |
0.12 |
>32 |
32 |
8 |
8 |
>32 |
8 |
8 |
32 |
0.25 |
8 |
BC20/08 |
>32 |
>32 |
>32 |
>32 |
>32 |
8 |
8 |
8 |
>32 |
>32 |
32 |
32 |
>8 |
8 |
BC22/08 |
32 |
>32 |
1 |
0.25 |
>32 |
32 |
8 |
8 |
>32 |
8 |
8 |
32 |
>8 |
8 |
BC23/08 |
32 |
>32 |
2 |
0.5 |
>32 |
32 |
32 |
8 |
32 |
8 |
8 |
32 |
8 |
>8 |
Min |
1 |
>32 |
0.12 |
0.12 |
0.12 |
8 |
2 |
2 |
8 |
0.5 |
1 |
2 |
0.25 |
0.25 |
Max |
>32 |
>32 |
>32 |
>32 |
>32 |
32 |
32 |
8 |
>32 |
>32 |
32 |
32 |
>8 |
>8 |
MIC50 |
32 |
>32 |
1 |
0.25 |
>32 |
32 |
8 |
8 |
32 |
8 |
8 |
32 |
0.25 |
8 |
CHL: Chloramphenicol, CIP: Ciprofloxacin, CLI: Clindamycin, DFX: Danofloxacin, ENR: Enrofloxacin, ERY: Erythromycin, FLO: Florfenicol, LCM: Lincomycin, MBF: Marbofloxacin, OTC: Oxytetracycline, SPT: Spectinomycin, TIL: Tilmicosin, TUL: Tulathromycin, TYL: Tylosin
Guidelines: <2 µg/ml sensitive; >2 to <8 intermediate sensitivity; >8 resistant
Viral antigen detection
In the test for BVDV antigen, 18 % (2 cases) of the acute pneumonic samples 18 % (2 cases) of chronic pneumonic samples were found positive. None of adenovirus 3, PI, BRSV and BoHV1 antigens were detected in any of the samples.
Molecular typing of M.bovis
The results show the existence of two major clusters (Figure 1): the first (A) containing the majority of isolates (85 %) and the second (B) containing the remainder (15 %). The major cluster could be subdivided into two further groups. No relationship was found between genotypes and whether calves had died of acute or chronic pneumonia. Calves on three farms were infected with both cluster A strains and B strains.
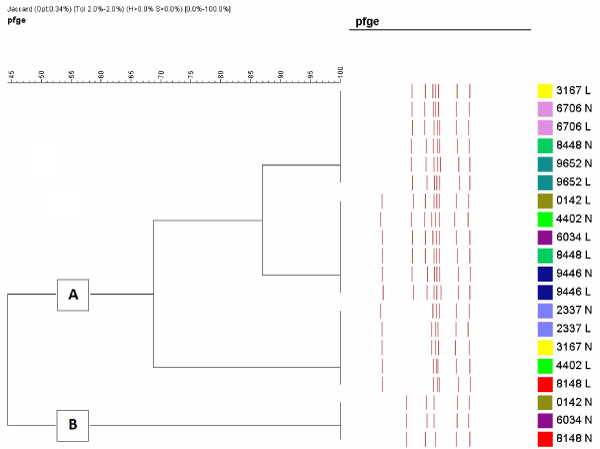
Figure 1. Molecular typing of Mycoplasma bovis strains isolated from calves with pneumonia showing two distinct clusters. Strains with the identical colour are from the same calf.
It was previously reported that M. bovis and BVDV play an important role in chronic pneumonias in feedlot cattle [17]. In this study, the percentage of BVDV antigen positivity in acute and chronic pneumonic samples were found to be the same. None of adenovirus 3, PI, BRSV and BoHV1 antigens were detected in any of the samples. It is clear from this study that M. bovis is the major pathogen in this feedlot system being isolated from both acute and chronic lesions of calves dying of respiratory disease. It is probably also widespread throughout the cattle industry in Turkey [5] as it is in many countries worldwide [18].
The results described here confirm those in other studies [6,7] which shows the lack of in vitro susceptibility of many antibiotics against M. bovis including tylosin, tilmicosin, erythromycin, chloramphenicol and ciprofloxacin and partial susceptibility to florfenicol, spectinomycin, danofloxacin, enrofloxacin and marbofloxacin. Antibiotics which still appear effective in vitro comprise lincomycin, clindamycin and tulathromycin. The results presented here are of great concern particularly that evidence of resistance to the fluoroquinolones is being seen which is alarming for human health where these antibiotics are often the last option for treatment of resistant infections [18].
While all isolates of both M. haemolytica and P. multocida were resistant to gentamicin, some P. multocida isolates were resistant to erythromycin and tylosin, trimethoprim-sulphametoxasol and tilmicosin, tulathromycinto enrofloxacin and tetracycline. M. haemolytica isolates were largely resistant to erythromycin, tylosin, tetracycline, trimethoprim-sulphametoxasol, but less so to tilmicosin, enrofloxacin, marbofloxacin, florfenicol, ampicillin and penicillin.
When the occurrence and spread of resistance in Pasteurella, Mannheimia and M. bovis organisms is better documented at herd level, this information will help bovine practitioners to minimize the number of therapy failures of bovine pasteurellosis related to antimicrobial resistance.
The results of the molecular typing confirm those of McAuliffe et al [15] who reported the existence of two markedly distinct groups by several molecular typing tools suggesting two divergent lines of descent that arose during evolution, one from north America and another from Europe. It seems that the European M. bovis strains may have mutated from the very closely related M. agalactiae which is a pathogen of sheep and goats [15]. It would have been interesting to know the clinical history of the farms where strains of both clusters were detected but these were not recorded. It could be expected that mortality and morbidity may have been higher as mixed strains sometimes give rise to exacerbation of disease [18].
The work reported here indicates that vaccines are urgently needed to combat these infections as antibiotics are clearly failing. However it will be necessary to show that the vaccines are protective against the two major clusters.
- Catry B (2005) Pasteurella and Mannheimia species from calves: differentiation and antimicrobial resistance. Faculty of Veterinary Medicine, Ghent University, b-9820.
- Nicholas RA, Ayling RD (2003) Mycoplasma bovis: disease, diagnosis, and control. Res Vet Sci 74: 105-112. [Crossref]
- Gagea MI, Bateman KG, Shanahan RA, Dreumel T, McEwen BJ, et al. (2006) Naturally occurring Mycoplasma bovis–associated pneumonia and polyarthritis in feedlot beef calves. J Vet Diagn Invest 18: 29-40. [Crossref]
- Özdemir Ü, Erdenlig S (2006) Dot-immunobinding technique for the identification of Mycoplasma species isolated from ruminants. Proceedings of VII National Veterinary Microbiology Congress. 26-28, p119.
- Sayin Z, Sakmanoglu A, Uçan US, Uslu A, Hadimli HH, et al. (2016) Mycoplasma infections in dairy cattle farms in Turkey. Turk J Vet Anim Sci 40: 569-574.
- Ayling RD, Baker SE, Peek ML, Simon AJ, Nicholas RAJ (2000) Comparison of in vitro activity of danofloxacin, florfenicol, oxytetracycline, spectinomycine and tilmicosin against recent field isolates of Mycoplasma bovis. Vet Rec 146: 745-747. [Crossref]
- Barberio A, Flaminio B, De Vliegher S, Supré, K, Kromker, V, et al. (2016) In vitro antimicrobial susceptibility of Mycoplasma bovis isolates identified in milk from dairy cattle in Belgium, Germany, and Italy. J Dairy Sci 99: 1-7. [Crossref]
- Nicholas RAJ, Ayling RD, Stipkovits L (2002) An experimental vaccine for calf pneumonia caused by Mycoplasma bovis. Vaccine 20: 3569-3575. [Crossref]
- Nicholas RAJ, Rosales RS, Loria GR (2017) Mycoplasma the big issues. Anim Husb Dairy Vet Sci 1: 1-4.
- McAuliffe L, Ellis R, Lawes J, Ayling RD, Nicholas RAJ (2005) 16S rDNA and DGGE: a single generic test for detecting and differentiating Mycoplasma species. J Med Microbiol 54: 1-9. [Crossref]
- Rosales RS, Churchward CP, Schnee C, Sachse K, Lysnyansky I, et al. (2015) Global MLST analysis of Mycoplasma bovis isolates reveals two main population clusters. J Clin Microbiol 53: 789-794. [Crossref]
- Koneman EW, Allen SD, Janda WM, Schreckenberger PC, Winn WC (1992) Haemophilus. In, Koneman EW (Ed): Color Atlas and Textbook of Diagnostic Microbiology. 4th Ed, 288-292, Lippincott Company.
- Nicholas RAJ, Baker SE (1998) Recovery of mycoplasma from animals. In, Miles RJ, Nicholas RAJ (Ed): Mycoplasma Protocols. 37-44, Humana Press, Totowa, NJ,
- Poveda JB, Nicholas RAJ (1998) Serological identification of mycoplasma by growth inhibition and metabolism inhibition test. In, Miles RJ, Nicholas RAJ (Ed): Mycoplasma Protocols. 105-112, Humana Press, Totowa, NJ.
- McAuliffe L, Kokotovic B, Ayling RD, Nicholas RAJ (2004) Molecular epidemiological analysis of Mycoplasma bovis isolates from the United Kingdom shows two genetically distinct clusters. J Clin Microbiol 42: 4556-4565. [Crossref]
- Bauer AW, Kirby WM, Sherris JC, Truck M (1966) Antibiotic susceptibility testing by a standardized single method. Am J Clin Pathol 45: 493-496. [Crossref]
- Shahriar FM, Clark EG, Janzen E, West K, Wobeser G (2002) Coinfection with bovine viral diarrhea virus and Mycoplasma bovis in feedlot cattle with chronic pneumonia. Can Vet J 43: 863-868. [Crossref]
- Nicholas RA (2011) Bovine mycoplasmosis: silent and deadly. Vet Rec 168: 459-462. [Crossref]

