Mycoplasma diseases of livestock such as contagious bovine pleuropneumonia, contagious caprine pleuropneumonia and contagious agalactia still represent major problems for animal health authorities worldwide. Many significant improvements have been seen as a result of research into these degenerate bacteria, mainly in the area of diagnosis, with the availability of specific, effective new molecular tools to detect small quantities of these pathogens (some of them highly fastidious, unculturable and often in mixed bacterial culture) directly from clinical samples. The objective of the present study was to identify and differentiate Mycoplasma species of sheep, goats and cattle by a new diagnostic test based on PCR of the 16S rRNA gene with specific primers and separation of the PCR product according to primary sequence using denaturing gradient gel electrophoresis (DGGE). Detection and identification results were compared with the ones obtained by sequence analysis. The development of a PCR DGGE offers advantages of a rapid identification of many Mycoplasma species for which no specific PCR is yet available, enabling the differentiation of animal Mycoplasma species. It represents a significant improvement from conventional culture and serological tests in term of specificity and time to perform.
colorectal cancer, complication, estimated blood loss, length of hospital stay, network meta-analysis, operation time
Mycoplasmas cause a wide range of disease in both humans and animals and are commonly associated with pneumonia, arthritis, conjunctivitis, infertility and abortion. Mycoplasmas are highly fastidious, typically taking weeks to culture while many serological tests are non-specific and poorly sensitive. More recently PCRs have been used to detect Mycoplasma species, but it is not feasible to develop PCR tests for each species. Consequently, there is a significant need for a single, generic test that can both detect and differentiate Mycoplasmas. Denaturing gradient gel electrophoresis (DGGE) can theoretically detect single base mutations in DNA [1-3]. Amplicons of the same length but with different nucleotide compositions are separated in a denaturing gradient gel of polyacrylamide, based on their differential denaturation profile. During denaturation, the two strands of a DNA molecule separate or melt at a specific denaturant concentration, and the DNA sequence stops its migration in the gel. PCR DGGE allows the detection and differentiation of mixed and very small amounts of Mycoplasmas showing higher sensitivity than the conventional tests. Using this method McAuliffe et al. [4,5] enabled the differentiation of 67 Mycoplasma species of human and animal origin representing a significant improvement on current tests. Diagnosis of Mycoplasma infection can be made directly from clinical samples in less than 24 h and can even detect new and unculturable Mycoplasmas. We report the application of DGGE to detect pathogenic and nonpathogenic Mycoplasmas from small ruminants
DNA was extracted from a 1ml aliquot of stationary-phase culture using the InstaGene Matrix kit according to the manufacturer’s instructions (Bio-Rad) and from swabs and tissues using the Genomic DNA GenElute kit according the manufacturer’s instructions (Sigma). A total of 75 clinical samples from a wide variety of ruminant diagnostic samples including nasal, eye, ear swabs, lung tissue, milk, brain tissue, synovial joint fluid were analysed.
Amplification of the V3 region of the 16S RNA gene was performed by using universal bacterial primers GC-341F and the mollicutes-specific primer 534R [5] and the AmpliTaqGoldDNA Polymerase kit (Applied Biosystems) according to the manufacturer’s instructions. Aliquots were checked for correct amplification by electrophoresis on a 2% agarose gel followed by visualization with GelRedTMNucleic Acid Gel Stain under UV illumination.
Mycoplasma DNA was successfully amplified for DGGE, performed using the DCode™ Universal Mutation System (Biorad). Samples (20 ul) were loaded on to 10% polyacyrlamide/bis (30:1) gels with denaturing gradients from 30 to 60%, in 1 × TAE electrophoresis buffer (Severn Biotech). Electrophoresis was performed at 100V at a temperature of 60°C for 18-20 h. Gels were then stained with SBYR Gold (Cambridge BioScience,) in 1 × TAE for 30 min at room temperature and visualized under UV illumination.
The identification was confirmed by sequence analysis of 16S rRNA gene, and the genomic sequences were downloaded from the National Center for Biotechnology Information (NCBI) resource (http:// www.ncbi.nlm.nih.gov). Sequencing and BLAST revealed the taxonomic informations.
In total 54 clinical samples were successfully examined by DGGE and found to be positive for Mycoplasma infection. All Mycoplasma species tested produced a PCR product of approximately 340 bp (Figure 1).
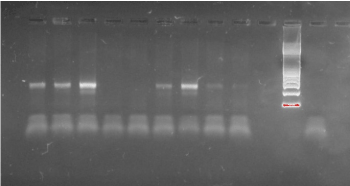
Figure 1. Electrophoresis of PCR products. All Mycoplasma species amplified a 340 bp amplicon
The use of DGGE to separate PCR products enabled differentiation of the different Mycoplasmas species analyzed (Figures 2-3). This indicated that the sequence was different for each isolate because the migration of the bands in the gels is a function of the melting behaviour of the amplicons in the chemical gradient used.
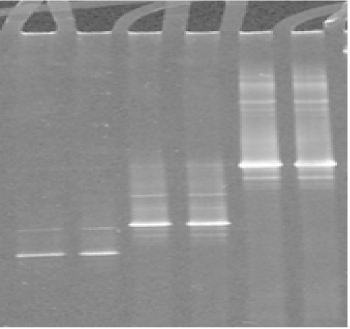
Figure 2. DGGE analysis of the amplified V3 region of 16S rRNA gene from ruminant mycoplasma type strains: Lanes 1, 2 : M. agalactiae; lanes 3, 4: M. bovis; lanes 5, 6: M. mycoides subsp capri
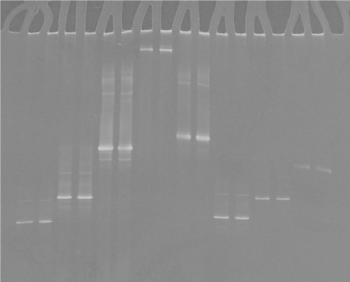
Figure 3. DGGE of ovine, caprine, bovine and avian mycoplasma controls: lanes 1, 2 : M. agalactiae, lanes 3, 4: M. bovis; lanes 5, 6: M. mycoides, lanes 7, 8: M. gallisepticum, lanes 9, 10: M. synoviae,, lanes 11, 12: M. gallinaceum, lanes 11, 12: M. gallinarum, lanes 13, 14: M. meleagridis.
The presence of multiple bands indicated that more than one 16S rDNA operon was present and that there were some sequence differences between the copies. Our experience revealed some samples contained a multiple infection of M. m. capri and M. yeatsii on a Sicilian farm (Figure 4). The sequences were submitted to the EMBL Data Library according to the EBI Procedure under the accession number CP010267.1 and JN935862.1. A brain sample giving a pattern band typical for M. arginini was submitted under the accession number JQ903581.1.
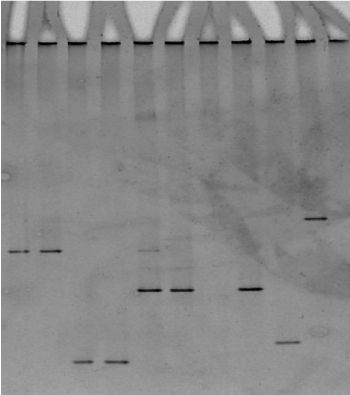
Figure 4. Presence of different bands indicating multiple infection. Lanes 1, 2: M. mycoides, lanes 3, 4: M. agalactiae, lanes 5, 6 multiple bands, (M. mycoides, M. yeatsii), lane 7 M. arginini, lane 8: M. yeatsii.
Sequencing the 16S rDNA amplified from genomic DNA giving a DGGE pattern specific for M. m. capri (Figure 5 lanes 7,8) and comparison with the equivalent 340 bp amplicon derived from a reference M. capricolum subsp. capricolum sample revealed 100% sequence coverage (accession number AF086703).
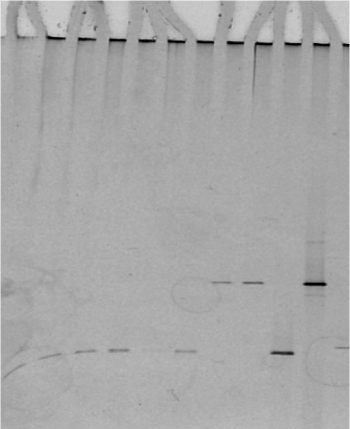
Figure 5. DGGE of Mycoplasma species tested: lanes 1-8 unknown mycoplasma samples confirmed with DGGE, lane 9: M. agalactiae control, lane 10: M. mycoides control
DGGE enabled the rapid detection and differentiation of Mycoplasma species and can be used to diagnose infections either directly from tissues or from cultured isolates. This method allows the identification of over 65 Mycoplasma species in a single test and in a lesser expensive way than other fingerprinting techniques. Furthermore, it is capable of detecting Mycoplasmas in mixed cultures (which would have been difficult to detect by conventional methods) and/or new mollicute species and is suitable for routine use in the diagnostic laboratory.
A major advantage of the DGGE is its potential to visually profile and monitor changes occurring in various microbial communities that are undergoing different treatments or modifications [6]. In addition, a great advantage of DGGE is that it is possible to obtain taxonomic information because bands can be excised, re-amplified and identified by 16S rDNA sequencing. Using this technique, diagnosis of emerging infections could become easier and faster, and identification of uncultivable pathogens can also now be facilitated.
- Fisher SG, Lerman LS (1983) DNA fragments differing by single base-pair substitutions are separated in denaturing gradient gels: correspondence with theory. Proc Natl Acad Sci U S A 80, 1579-1583. [Crossref]
- Garcia M, Jackwood MW, Head M, Levisohn S, Kleven SH (1996) Use of species-specific oligonucleotide probes to detect Mycoplasma gallisepticum, M. synoviae, and M. iowae PCR amplification products. J Vet Diagn Invest 8: 56-63. [Crossref]
- Lerman LS, Beldjord C (1999) Comprehensive mutation detection with denaturing gradient gel electrophoresis. In: Cotton, R.G.H., Edkins, E. and Forrest, S. (Eds) Mutation Detection. Oxford University Press, Inc., New York, 35-61.
- McAuliffe L, Ellis R, Ayling RD, Nicholas RAJ (2003) Differentiation of Mycoplasma Species by 16S Ribosomal DNA PCR and Denaturing Gradient Gel Electrophoresis Fingerprinting. J Clin Microbiol 41: 4844-4847. [Crossref]
- McAuliffe L, Ellis RJ, Lawes JR, Ayling RD, Nicholas RAJ (2005) 16S rDNA PCR and denaturing gradient gel electrophoresis; a single generic test for detecting and differentiating Mycoplasma species. J Med Microbiol 54: 731-739. [Crossref]
- Muyzer G, de Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59: 695-700. [Crossref]





