Abstract
There are many evidence in scientific literature that relate to causal nutrition and risk of disease. The concept that proper nutrition can be a significant protective factor that can significantly affect the risk of cardiovascular and neoplastic diseases is widely demonstrated by scientific literature. Evidence has been shown at an epidemiological level first: it has been shown that a certain qualitative and quantitative composition of nutritional support can be both a protective factor that can reduce the incidence of pathologies and, in other dietary formulations, a a causal factor that can increase its etiopathogenetic effect. It is well known, for example, how the incidence of cardiovascular disease is closely related to the lifestyle of the person, and in particular the excess energy and/or lipids, particularly saturated. Likewise, the characteristics of the diet also significantly affect the incidence of neoplastic diseases. It is believed that on the combination of neoplastic and cardiovascular diseases, which then represent the first two causes of death in the most developed countries, at least one-third of case histories recognize diet as the main causal factor. While the first evidence of nutrition and health relationship has come from epidemiological data on populations, subsequent research is progressively identifying the molecular pathophysiological mechanisms at the basis of the observed effects. The evidence that they are showing shows significant mechanisms involved in nutrition. Nutrients, ie molecules absorbed within the body by the digestive tract, are classically divided into energetic and structural, wanting to indicate with what food-derived molecules can or give the energy needed for various metabolic processes or become part of the cellular and tissue structures of the body. The study of the relationship between nutrition and health has recently added to those categories that of chemiopreventive or bioactive nutrients. With this term we want to point out those molecules made by the diet, for which, regardless of their role as nutritional energy or structural, it is noticed how the magnitude of their representation in the diet is associated with a reduction in the risk of pathologies, sometimes being also known explanations of the molecular mechanism with which this occurs. For example, polyunsaturated omega-3 fatty acids derived from diet seem to be the basis of the mechanism for which epidemiology has shown that a diet with various weekly fish portions significantly reduces cardiovascular risk. Many other substances, often with protective power against the mechanisms of oxidative cell damage, are increasingly placed under careful attention to explain the relationship between nutrition and health at the molecular level.
Key words
bioactive nutrients, chemoprotective nutrients, health, nutrition, nutrigenomics, oxidative cell damage
Introduction
It is known that a diet rich in fruit and vegetables represents a remarkable tool for preventing the primary pathology not only by providing optimally energy and relative shares of carbohydrates, proteins and lipids, but also because of the nutrients that are derived from foods bioactive, even in small quantities but with important biological effects such as polyphenols, soluble and insoluble fiber, polyunsaturated fatty acids, etc. [1-5]. The nutrition status of a subject, in addition to the clinical evaluation of energy balance analysis, body composition and functional changes, within the parameters of general nutrition diagnostics, should also be considered in relation to the evaluation of 'exposure to bioactive nutrients and their consequent effects, in particular as regards the effects on the risk of subsequent pathologies, also in reference to far-reaching temporal evolution. Traditionally, nutrition assessment has always been aimed at diagnosing malnutrition, often understood as being defective or, more commonly, in Italian society, in excess of energy or even in the absence of specific nutrients. Notwithstanding the importance and validity of this approach, but also considering the enormous growth data of the incidence of obesity, it should also be considered that from further evaluation of the state of nutrition it is possible to obtain other relevant information [5-10]. In this sense, the molecular diagnosis of the state of nutrition in an individual requires fine information on the presence in biological fluids and tissues of bioactive nutrients and their metabolic effects in order to assess how and to what extent their nutrition its long-term risk of cardiovascular and neoplastic diseases. Additionally, individual genomic variables that can influence the effects of bioactive nutrients by regulating, absorbing, metabolizing, excreting, and the quality and quantity of biological effects will also need to be considered [9-15].
Nutrigenomics
Nutrigenomics will therefore be an individual genomic analysis aimed at examining the genome and its expression in relation to the effects of nutrients derived from the diet. The different individual susceptibility to the effect of nutrients can however result in the fact that the effect of diet on gene expression is partly subject to variability [16-22].
Figure 1 wants to schematically represent how the relationship between exposure to bioactive nutrients in relation to the state of nutrition, which however concerns the development of nutritional situations in the long term and not individual moments, comes to affect the health of an individual, acting on the magnitude of the risk of disease. This effect, however, is carried out through a series of mechanisms that can greatly affect the characteristics. The bioactive nutrient can be variable absorbed as well as subsequently variably activated or inactivated. His biological activity will thus expose on specific molecular "targets" which may also be variable or modulable in relation to the genomic characteristics of the individual affecting its quality and quantity [22-25].
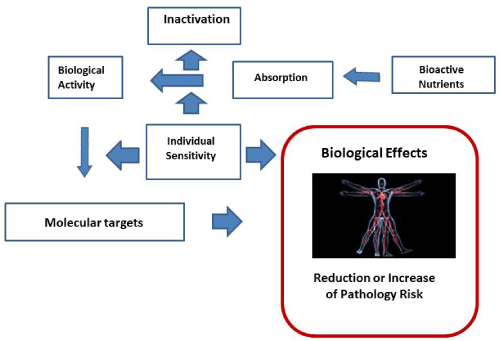
Figure 1. Relationship between exposure to bioactive nutrients and biological effects in the individual
The availability of objective tools to evaluate the presence and effects of bioactive nutrients in a single patient therefore becomes a diagnostic procedure, that is to identify a pathology and defining its condition. Naturally, at present, diagnostics can only cover nutrients the effects of which are already known, while progress in research will increasingly present more in-depth studies: for example, only a few tens of flavonoids, bioactive nutrients of food origin, which have nutritional data deepened, while it is estimated that an ordinary diet will bring thousands. A bioactive nutrient or a metabolite produced in relation to it whose analysis produces valid diagnostic indications can therefore be termed nutrition biomarkers. Similarly, they can act as nutritional biomarkers, in addition to nutrient metabolites, including gene expression profiles produced by them.
More and more, in fact, technology allows multiple analysis of the genome set activity. Consequently, it is now possible, within nutrigenomics, to analyze changes in the expression of an entire genome, both at RNA level messengers and protein products, in relation to specific nutrition conditions. In this way it becomes possible to evaluate the effects of nutrition even without knowing the individual nutrients that are responsible for it but evaluating its effect as a whole. This is of particular importance when considering how in many cases the biological effects of various nutrients are different according to whether they are tested individually or in combination [26-32].
Molecular diagnosis of the state of nutrition
The objective of the molecular diagnosis of the state of nutrition will therefore be to evaluate in the individual subject, on the basis of the objective use of nutritional biomarkers, the risk of pathology related to dietary characteristics. At a time when it is possible to do this in previous situations, the clinical manifestations of the disease is then defined as a secondary prevention intervention capable of allowing the formulation of a diagnosis of a disease at an early and preclinical stage. Early diagnosis in this case, but not only, also corresponds to more effective intervention opportunities; however epidemiological data show that positive modification of nutritional behaviors will have beneficial effects at any stage of the development of a related pathology [32-44].
The diagnostic definition of the state of nutrition in general clinical practice is primarily aimed at assessing qualitative and/or quantitative malnutrition. Of the three levels in which nutrition status analysis is generally developed, which is the evaluation of energy balance, body composition evaluation, and evaluation of related functional modifications, the first is used most since it has already been sufficient responses to guide the diagnostic process. The energy balance can be evaluated by comparing contributions estimated by an anamnestic interview to energy consumption measured with appropriate instrumentation. With the nutrition interview, it is possible to collect information on the quality and quantity of the foods made, and then formulate a rough, naturally approximate assessment of the dietary complex. With instrumental evaluations it is possible to measure the energy consumption at rest and also in controlled exercise situations. However, the anthropometric data is often sufficient: height, body weight, body mass index (weight ratio in kg/height in m high per square), fat mass percentages and lean mass (measured by impedance measurement). Table 1 shows the main anthropometric and bioumoral parameters used in the general evaluation of nutrition status [40-53].
Table 1. General nutrition status evaluation indices
Anthropometric indices |
Body mass |
It typically grows in relation to excess energy input. Values to be compared to peoples comparable to age and sex of the subject |
Height |
Dependent on growth and skeletal length. Values to be compared to peoples comparable to age and sex of the subject |
IMC o BMI, body mass index |
It allows to combine in a single weight value and height (calculated from the weight ratio in kg / height in m high per square). Values to be compared to peoples comparable to age and sex of the subject |
Mass Percentage lipid |
It typically grows in relation to excess energy input. It can be evaluated by measuring the thickness of skin patches in the reference anatomical points, electrical impedance measurements that grow to increase the proportion of adipose tissue, body density measurements and dilution of isotopes. |
Abdominal circumference |
It typically grows in relation to excess energy input. It is one of the key criteria for formulating the diagnosis of metabolic syndrome |
Biochemical Indexes |
Triglycerides |
Dependent on nutrition and also by individual factors. Increased cardiovascular risk with increasing its concentration. |
Cholesterol |
Dependent on nutrition and also by individual factors. Increased cardiovascular risk with increasing its concentration. |
HDL-cholesterol |
Dependent on nutrition and also by individual factors. Increased cardiovascular risk with reducing its concentration. |
LDL-cholesterol |
Dependent on nutrition and also by individual factors. Increased cardiovascular risk with increasing its concentration. |
The set of parameters in Table 1 allows for first analysis, in the context of nutritional clinical assessment, to measure the degree of adiposity of a subject and the main lipidemic indexes to carry out a nutrition assessment, especially in terms of amount of energy, but also about the quality of the diet composition observed. Excessive energy intake is the most common cause of obesity, although many other etiologies need to be considered. Similarly to dyslipidemia, however, in their various aspects are an important risk factor for cardiovascular disease. A classic nutritional deficiency index is hypoalbuminemia, particularly in elderly subjects. Other laboratory indices useful in this regard are quantization of transferrin and lymphocytes in the blood [40-57] (Table 2).
Table 2. Assessment indexes of general nutritional deficiency
|
DEFICIT |
Mild |
Moderate |
Serious |
Albumin (g/dl) |
3.5-3.2 |
3.2–2.8 |
<2,8 |
Transferrin (mg/dl) |
200-180 |
180-160 |
<160 |
Lymphocytes (n./mm3) |
1800-1500 |
1500-900 |
<900 |
While these indices mainly concern the overall nutritional deficit and therefore, first of all, total energy input, other specific biomarkers are needed to diagnose specific nutrient deficiencies. In this sense, in many cases it is possible to diagnose a deficiency of a specific nutrient, such as a vitamin, prior to the development of clinical symptoms by dosing it in fluid or biological tissues. In other cases it is possible to evaluate a biological activity closely related to the presence of that nutrient: for example, the enzyme glutathione peroxidase of erythrocytes requires selenium and when this is deficient it decreases its activity. Similarly, it can be done for any other essential nutrition, that is to be obliged to be present in the diet to avoid symptoms of deficiency. These indices are therefore very useful as an objective reference of situations in which nutrition is not adequate, that is to provide the right amount of energy and nutrients [40-60]. Appropriate nutrition is defined as the one in which every essential nutrient is present at least to the recommended nutrition ratio (LARN, recommended daily amounts of nutrition). It is also necessary that the amounts of proteins, carbohydrates and lipids are appropriately distributed within the total energy quota, which must, however, also be appropriately related to the subject's characteristics. In addition, a number of nutrients must also be contained below certain quantities as they are toxic to excessive concentrations. When, however, with the satisfaction of these primary nutritional needs, we also want to monitor to what extent an individual with the diet is exposed to the activity of nutrients that can also influence the risk of disease and ultimately also the 'Life expectancy, it is necessary to use other and more fine molecular parameters [40-60]. The objective therefore becomes to have diagnostic references to indicate to what extent the diet has or exerts effects on the risk of disease. In this sense, the technique used must be more thorough and extending to multiple molecular effects analysis can be defined as a molecular diagnostic of the state of nutrition.
Various types of analysis are possible in this sense: they can be distinguished in two main categories: to measure the quality and quantity of nutritional benefits of bioactive nutrients and to measure their effect as well as individual sensitivity to them. In fact, the two aspects often overlap and can not always be discriminated against. Therefore, biomarkers can be distinguished by biomarker and biomarker of the effect of bioactive nutrients [40-60].
Biomarker intake
The direct dosing of bioactive nutrients in fluid and body tissues is certainly a diagnostic procedure quite independent of the individual's sensitivity to them. It is well known that the presence of bioactive nutrient serum, such as the various polyunsaturated fatty acids of the omega-3 and omega-6 series, is quite proportional to their representation in the diet prior to the withdrawal, although this is fasting, and therefore independent of the first intestinal absorption of nutrients. Of course, even in this case, there may be a variability of the individual in relation to the nutrition absorption efficiency which may vary in relation to the use of receptors and/or conveyors with variants of different activity and its ability to catabolize it . For many years, literature has shown that serum levels of nutrients such as vitamin A, vitamin E and selenium correlate significantly with the incidence of various forms of cancer. These substances, in addition to being the essential nutrients essential to the diet, have, at higher dosages, also the chemiopreventive effects. In the 1990s, several clinical epidemiology studies were carried out in the study of the effect of the prevention of neoplastic and cardiovascular diseases by the administration of vitamin A or beta-carotene supplements, which is its precursor [40-65].
Surprisingly, these studies had to be discontinued earlier than expected due to preliminary evidence of a potential increase in health risks due to administration. In fact, the logical basis of experiments was very solid and consistent with the observation that in populations where serum beta-carotene levels were higher, there was a significantly lower risk of neoplastic and cardiovascular disease. For this reason, it was thought that there was a causal relationship between the major serum beta-carotene concentrations and the reduction of risk. Evidently this has not been proven to be true and it is presumed that the reason for this is that, while excessive intake of vitamin A or beta-carotene may have toxic effects, they did not appear in nutrition benefit as in this case these nutrients were part of the nutrient complex made by a given diet, but it is not possible to distinguish at this level from which nutrients depend on the observed effects [57-70].
For example, it is well-known that a diet rich in fruit and vegetables has significant preventative effects, which is the indication of the world's leading health authorities, but also by designing characteristic features of the so-called Mediterranean Diet, that is, that whole of nutritional behaviors of populations of the Mediterranean area for which for the first time it has been possible, since the 50's, to demonstrate close relationships between nutrition and health. Surely those who feed on such a diet have higher beta-carotene serum concentrations, but apparently this substance is absorbed by foods along with many others, of which surely most must still be subject to characterization, some of which will have greater bioactivity and other minor. If this type of experimental evidence has highlighted the difficulty of transferring bioactive nutrient-derived observations in food-processing applications, they clearly highlighted the concept of different nutritionally-suited foods in the body. These concentrations can then be subjected to diagnostic analysis: if the simple serum concentration of beta-carotene is associated with a significant reduction in the risk of pathology in the populations, simultaneous analysis of a large number of nutrients in the single subject will be able to produce even more detailed information on the one hand in terms of objective assessment of the nutrition regime adopted, on the other in terms of the resulting risk entity configured [57-70].
This type of analysis, unlike interviews and nutritional questionnaires, allows you to have objective data on exposure to bioactive nutrients in the diet. It should also be considered that nutritional content of nutrients can only be estimated on the basis of medium compositions, but in some cases they are also subject to considerable variability: eg, the same type of food according to the origin can have enormous differences in selenium content, or the content of glucorafanine of the different broccoli varieties may also vary by 25 times. Although diagnostic analyzes of this kind are not yet in use, the set of observations available by scientific literature already allows for highly predictive serological determination of a complex of bioactive nutrients in a single patient. However, there is still a need to develop chemical analysis methods compatible with the operational needs of the clinical analysis laboratory, especially in terms of cost and reproducibility in large-scale use.
Among the parameters for which literature indicates ever greater and more indicative correlations with the nutrition status are saturated and unsaturated fatty acids derived from nutrition. Table 3 below shows the main doses in serum and their concentration varies with the amount of food intake that they contain [70-75].
Table 3. Fatty acids used as serum biomarkers of nutrition status
Fatty Acid |
Saturation |
Carbon atoms: Double bonds |
Myristic acid |
saturated |
14:0 |
Palmitic acid |
saturated |
16:0 |
Stearic Acid |
saturated |
18:0 |
Palmitoleic acid |
unsaturated omega-7 |
16:1 |
Oleic acid |
unsaturated omega-9 |
18:1 |
Linoleic acid |
unsaturated omega-6 |
18:2 |
Gamma-linolenic acid |
unsaturated omega-6 |
18:3 |
Alfa-linolenic acid |
unsaturated omega-3 |
18:3 |
Arachidonic acid |
unsaturated omega-6 |
20:4 |
Eicosapentaenoic acid |
unsaturated omega-3 |
20:5 |
Docosahexaenoic acid |
unsaturated omega-3 |
22:6 |
Table 3 shows the saturation degree and the number of carbon molecules of molecule length for each fatty acid. Saturated fatty acids, typically derived from the lipids of food of animal origin, are therefore more abundant in serum in relation to a higher representation of these products in food. Consequently, they are also associated with the risk of cardiovascular disease. Unsaturated fatty acids are instead derived from vegetable products and fish products. Various studies are accumulating evidence that the dosage of these substances in serum can be traced back to the quality of the lipid content of the diet. Moreover, plasma concentrations of the various types of fatty acids demonstrate significant predictive value of the risk of myocardial disease. Consequently, the serum dosage of the various types of fatty acids coming from different feedstock ratios relative to their presence in foods is an example of a valid reference nutrition biomarker. Obviously, further studies on even larger populations are still needed to identify the precise dosage modalities and significant reference values [70-75].
Biomarker effect
The effect of a biomarker indicates the presence and magnitude of a biological effect resulting from exposure to a nutrient. In practice it also measures the magnitude of a possible positive or negative effect on health. Cellular processes influenced by the bioactive components of foods are numerous, including, inter alia, cell proliferation and apoptosis, inflammation, differentiation, angiogenesis, DNA repair and carcinogen activation, ultimately entering fully in the complex of the etiopathogenetic phenomena of major cardiovascular and neoplastic diseases (Figure 2).
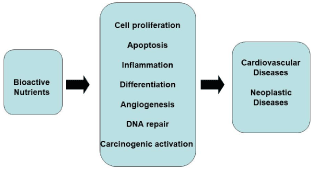
Figure 2. Influence of bioactive nutrients of foods on multiple biological processes whose alterations come into the pathogenesis of cardiovascular and neoplastic diseases
For all these processes, fundamental in maintaining physiological homeostatic equilibria, various bioactive nutrients are known to modulate its development. While it is still not possible to define the contribution of each nutrient to the various processes shown in the figure, it is evident that such a large spectrum of activity practiced by various nutrients with the complexity of their activities can easily explain how the quality of nutritional support may have a major influence on the health and the risk of illness. For example, among the nutrients that can induce cell apoptosis are quercetin, resveratrol and docosoesaenoic acid, all considered to be chemiopreventive nutrients. These effects are often described by analyzing the effect as pure substances on cultured cells. However, it is not said that the concentrations reached in vivo are sufficient to achieve the same effects, also considering the presence of other pro and anti-apoptotic factors. At present, there is sufficient evidence that only in some cases can be used to modify nutritional pathophysiological processes. Among these, various relate to biomarkers of lipid metabolism and adipose tissue activity [40-75].
Lipoprotein (a) (also called Lipoprotein LP (a) and Lp-a) is definitely a relevant diagnostic parameter in the assessment of the risk of cardiovascular disease, but also a biomarker of the effect of the state of nutrition as its levels however, influenced by the plasma. It is a subclass of lipoprotein whose concentration varies widely between different ethnicities, but also in relation to different pathological states such as coronary, cerebrovascular and atherosclerotic diseases. Different feeding modes, however, are able to adjust the level: for example, a diet rich in fish lipid derived products is able to lower it. In any case, desirable Lp (a) concentrations should be lower than 14 mg/dL, while a concentration of 14-30 mg/dL is considered "borderline", 31-50 mg/dL at high risk and higher than 50 mg/dL at high risk [40-75].
Adipocytes, hormone mediators produced by adipose tissue, leptin and adiponectin, as well as Apolipoprotein AI (ApoAI) and ApoB are other substances whose serum concentration varies in relation to the state of nutrition and the risk of cardiovascular disease. In the case of apolipoproteins, the increase in the ratio of apoB/apoA1 is also considered as an indicative biomarker of greater risk as well as indicative of a causality correlated nutrition [40-75].
Serum homocysteine levels have been shown to have some correlation with cardiovascular risk but also be dependent on nutritional factors. Homocysteine is considered to be capable of causing vascular endothelial dysfunction with formation of free oxygen radicals, interfering with the vasodilating and antithrombotic function of nitric oxide (NO). Its increase is due to the lack of vitamins, in particular folic acid, vitamin B6 and vitamin B12, and causes an increase in cardiovascular disease [40-75].
Oxidative damage
Other parameters considered to be biomarkers of the state of nutrition are related to oxidative damage. The oxidative damage is caused by an imbalance between the production of reactive oxygen species (free radicals) and the body's ability to detoxify the reaction intermediates readily or easily repair the resulting damage. Many bioactive nutrients have antioxidant capacity and therefore may thus exercise their power of prevention of disease, although often prove more specific mechanisms for each individual substance. It is, however, shown that, in relation to the characteristics of nutritional support, the total oxidative capacity of the blood changes. In this sense it is possible to monitor the effect of the nutrition status basically: 1) with the measurement of individual antioxidants in the plasma; 2) Measuring the total oxidation/total antioxidant capacity of the plasma; 3) by measuring the oxidation of lipoproteins [50-80].
Overall, these various parameters above are documented by the literature as correlated, in relation to their modulation, to the state of nutrition. Nevertheless, the use of these parameters is not always possible in the decision-making process following the diagnostic formulation. This depends on the fact that although there may be a statistically significant correlation with each of them, or even combinations, the margins of variation are sometimes too small to discriminate different states in the individual. Other times, the differences observed in certain groups of subjects analyzed are related to groups with particular characteristics that do not necessarily reproduce in other situations. In practice, though there are numerous parameters that have biomarker characteristics of the nutrition status, it is not possible to fully satisfy that which is a consolidated diagnostic and prognostic requirement: the need to be able to trace within the individual subject, and not on an average of a population, in the past nutritional history, and nevertheless able to evaluate at a contingent moment the future prospect in terms of the risk of specific pathologies [50-80].
However, the usefulness of this information goes beyond the diagnostic formulation, which may also affect the different therapeutic options that are commonly used and potentially identifiable in relation to the best health assessment possibilities. Based on these considerations, it is evident that the best application prospects in nutrition diagnostics concern multiple analysis systems both in terms of systematic analysis of gene expression, but also in systematic quantization of large numbers of nutrients and/or their metabolites . In fact, the diagnostic capacity, instead of individual markers, on a large set of them, can only become more accurate and can provide more information that can have a much greater impact on clinical operation [50-80].
As a consequence of these goals, the disciplines of nutrigenetics and nutrigenomics were born. Nutrigenetics concerns issues related to how the individual's genetics, detectable on the basis of single nucleotide polymorphisms (SNPs), the number of genetic copies and epigenetic phenomena, can influence its susceptibility to the diet . Nutrigenomics takes care of how the diet affects genetic transcription, protein expression, and metabolism. The main problem with nutrigenomics is to integrate genomics, that is, the multiple analysis of the functioning of all genes, with transcriptism, that is, the analysis of the whole gene expression gene products, and metabolomics, that is, the analysis all the way to the metabolic profiles, up to the "healthy" phenotype, that is, in other words that you want to pursue the best for maintaining the state of health and for the optimal prevention of pathologies. Mechanisms analyzed at various levels integrate into complex interrelations. These interrelations are schematized in Figure 3.
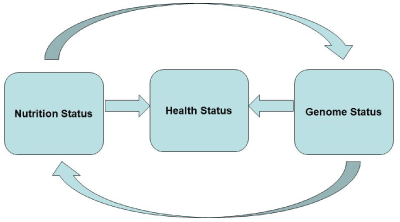
Figure 3. Interaction between nutrition and the genome in maintaining the health
It can be said that as many nutrients regulate the activity of many genes, the genome may still affect the activity of many nutrients, for example by modulating its bioavailability, absorption and, however, being able to be variable. The state of health, or illness, or predisposition to it also depends on these relationships. Genetics and genomics, applied in the context of nutrition and health, therefore have enormous potential for the development of biomarkers, indicators of predisposition to pathologies, knowledge of the effects of diet, even in the individual, as well as of individual bioactive nutrients [60-85].
The fact that the genome of many organisms has been sequenced makes it possible to predict with precision the respective transcripts, that is, the set of coded protein products. The multiple analysis of RNA messenger expresses, using the "DNA microchips" methodologies, allows to have a complete picture of the activity of each gene at a given time in a certain cell or tissue. At least at the present time, this degree of completeness can not be achieved neither with proteomics nor with metabolomics. For this reason, transcriptomycosis is generally the main study in differential regulation, including nutritional studies, to provide a framework for the metabolomic and proteomic analysis. Differential studies compare the differences between two multiple analyzes that compare two situations with a definite difference, for example in a cell such as RNA messengers are different in quantity, increased or decreased, compared to exposure or non-exposure to a given nutrient [50-80].
Proteomics
The recent development of proteomics is bound to the development of mass spectrometry technologies, which allow the analysis of complex protein mixtures at a level on one side and on the other, however, global. The main and most effective methods of mass spectrometry for these purposes are the electrospray ionisation (ESI) and the "matrixassisted laser desorption/ionisation" (MALDI). Using these methods, it is possible to compare protein blends and identify those that differ in different conditions. Molecules can be characterized by both the molecular weight and the specificity of fragments that can be obtained, based on the specific amino acid sequence of the protein, which can also be obtained from genomic and proteomic databases. However, proteomic studies are perhaps more complex than genomic [50-80]. This is not only because, in any case, genomic analysis technologies are still more powerful, but also because it is estimated that the number of proteins expressed by human cells as a whole is higher. At present, it is estimated that the number of human genes is about 25,000, while that of proteins of about 100,000, thanks to the splicing mechanisms of RNAs and the various post-translational modifications that allow to produce more proteins from the same gene. The proteomic study should be able to extend to three levels: expression, structuring and protein function. In fact in most cases, only the first level is practicable on a global scale with current technologies, while analysis of structure and function can only be made on smaller subgroups of proteins [50-80].
Transcriptomics
Studies on gene activity and regulation by "DNA microchips" are allowing enormous advances in the molecular understanding of disease mechanisms. Still more complex is to define the molecular basis of the state of health, as it is much less defined, even in general terms, than that of disease. On the other hand, global gene expression studies are beginning to contribute to this, especially in the nutrition field [50-80]. Studies on the expression of individual genes have also allowed and allow you to obtain important information about the effect on them of individual nutrients or exposure to a certain diet. Many genes have been shown to be regulated by various nutritional originators. This in some cases is directly related to the specificity of individual nutrients, for example fatty acids that modulate the expression of genes encoding lipid metabolism enzymes, but in other cases it has revealed less easily predictable mechanisms. In any case, it is extremely important that studies on individual and defined genes can coexist with the systematic and global study of all of them. This does not require starting from "candidate" genes that are believed to be nutritionally possible, but by working on all of these, it allows to highlight the complete picture of the affected genes, regardless of the availability of previous information about them [50-80]. The transcript of multiple transcription technology is one that uses "DNA chips". They are solid supports on which tens of thousands of different DNA probes are bound, and it is possible to test for the hybridization of the expression of as many genes. These "expression profiles" that are obtained "capture" the relative amounts of a large number of messenger RNAs, each in quantity related to the expression of the related gene. The comparison between two expression profiles obtained from two different but similar conditions makes it possible to identify the differences. Adjusting gene expression by nutrition interventions is often difficult to discriminate and interpret, because of the many but small variations that occur. Many genes whose gene product is of reduced abundance could be limited to a limited extent with signals to the capacity limits of the analytical technology. Consequently, differential expression studies related to nutrition require even more than in other cases the maximum standardization and calibration of the method to provide useful results [50-80].
Metabolomics
The most used metabolic profiling strategies are nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry (MS). Through these techniques it is possible to analyze the complex of metabolites present in a biological sample of nutritional interest, both qualitatively and with some limitations also related to sensitivity, at quantitative level. With the boundaries related to the methodologies, including those related to interindividual variability, they were determined in various metabolic profile researches related to various situations of nutritional interest.
The nutrigenomic technology complex has been used to search for nutritional biomarkers or related pathologies. Among these are in particular obesity and diabetes with which up to now it can be estimated that about 100 genes have been related. Of these, about twenty were specifically related to the diet. On the other hand, the particular increase that has been occurring in the Western world for about 20 years for these pathologies can certainly not be attributed to hereditary genetic alterations, but rather to the spread of incorrect food lifestyles. Various studies have also addressed the effect on the gene expression of the amount of energy involved as well as the extent of its protein, carbohydrate, and lipid proportions [50-87]. Other studies specifically focused on individual nutrients or combinations thereof. Many studies have focused on lipids, due to the importance of their role in the development of pathologies. In particular, the nutrigenomic effect of the degree of unsaturation and also the polyinsaturation of fatty acids derived from food as well as individual nutrients, such as docosoesaenoic acid, have been observed. Though the results are still too preliminary to be able to formulate specific indications to use this information to have expression biomarkers, some genes begin to be particularly alert about the effect of diet lipids. Among these are transcription factors "peroxisome proliferator activated receptors" (PPAR, alpha, beta and gamma), "liver X receptors" (LXRs, alpha and beta), and "hepatic nuclear factor-4 (HNF- 4) alpha ". Interestingly, "pyruvate dehydrogenase kinase-4 (PDK-4)" and "adipocyte differentiation related protein" (ADRP) have been found to be of interest to the PPAR receptor subcontracting. ipolipidemic drugs. Another transcription factor whose modulation depends on the contribution of lipids, but more specifically by their saturation is the SREBP-1 protein [50-87].Overall, many of these regulated genes have in common the fact of being bound by their binding regulatory regions of "heterodimeric nuclear receptors", ie formed by two different subunits. It is a class of proteins that bind DNA (DNA binding proteins) with regulatory function and in some respects similar to glucocorticoid and estrogen receptors, but are homodimeric, that is, formed by two equal subunits. The role of heterodimeric nuclear receptors is therefore of particular importance and on the other hand they are definitely involved in the etiopathogenesis of cardiovascular pathologies. It is interesting to note that two nutrients with significant evidence of chemopreventive capacity, such as docosoesaenoic acid and vitamin E, are also capable of regulating the action of various genes, including various regulated by diet lipids, via heterodimeric nuclear receptors . Among these genes can be remembered the one that encodes the enzyme "UDP-glucuronosyltransferase 1A1 (UGT1A1), which is responsible for bilirubin glucuronation and numerous xenobiotics. It is also interesting to note that the two monomers that together form the heterodimeric nuclear receptors one is usually the "retinoid X receptor" (RXR) whose typical ligand, retinol or vitamin A is also of nutritional origin. an example of genes and corresponding protein results regulated by fatty acids with decreasing (down-regulation) or up-regulation [50-87] (Table 4).
Table 4. Proteins produced by gene responses regulated by fatty acids and corresponding function
Up-Regulation |
Glut 4 |
Glucose Transport |
Pyruvate Kinase |
Glycolysis |
ATP Citrate Lyase |
Lipogenesis |
Glucose 6 Phosphatase |
Gluconeogenesis |
Fatty Acid Synthase |
Lipolysis |
Spot 14 |
Lipogenesis |
Stearoil.-Coa Denaturasi 1 |
Fatty Acid Desaturation |
Leptin |
Hormonal |
Down-Regulation |
Fatty Acid Translocase (FAT-CD36) |
Membrane Transport |
Fatty Acids Binding Protein (FABP) |
Intracellular Transport |
Lipoprotein Lipase |
Hydrolysis Triglycerides |
Acil-Coa Synthetase |
Activation |
Acil-Coa Oxidase |
Oxidation Peroxisomal |
Carnitine Palmitoyl Transferases 1 |
Activation Beta Oxidation |
Cytochrome P450A2 |
Microsomal Oxidation |
Phospho Enol Pyruvate Carboxykinase |
Gliceroneogenesi |
Uncoupling Protein 2 And 3 (UCP2, UCP3) |
Energy Production |
It can be seen how the genes in the table are referred in many cases to metabolic steps associated with the use of fatty acids as a source of energy. In many other cases, diet-modulated genes also affect other functions. A limitation of many of these data is that they have been obtained using bioptic tissues as samples for proteomic, transcriptional or metabolomic differential analyzes. Of course this is very diffi cult to propose in current clinical practice and in fact many data have been obtained at the animal level, with all the issues of interpretation of the validity of data also in humans. Obviously, further improvement in these technologies will have to give them a sense of sensibility and efficiency that can be used to make use of individual biological samples more easily accessible such as blood, urine or saliva [50-87].
The whole of possible or prospective future assessments of the state of nutrition can then be cross-checked with the diagnosis of the individual genetic profile. This, similarly to how it is proposed to become an important reference in many aspects of medical practice, may also be for the specific definition of individual susceptibility to various nutrients as well as certain characteristics of the diet as a whole. Various studies are going on in this regard and in the future one can assume that you can formulate "tailored" diets, that is customized for the individual. Having in-depth knowledge of molecular nutrition status, genes of susceptibility to the bioactive nutrient effects and consequent health effects, the diet proposed to the individual can be such as to maximize the beneficial effects [50-87].
Conclusions
Nutrigenomics is conceptually based on a number of considerations: 1) diet components can modulate gene expression; 2) Diet is a risk factor for various pathologies; 3) the expression of various genes modulated by the diet affects the risk of onset and progression of various pathologies; 4) The impact of a diet on health/disease balance may be affected by individual genetic factors; 5) nutritional interventions, even specific to the individual, may alter the development of various chronic conditions [88]. The main benefit of genomic analysis technologies in nutrition will enable the development of a series of diagnostic reference biomarkers based on proteomic, transcriptional and metabolomic profiles. For this to happen, it is not only necessary to collect information to diagnose and validate these biomarkers, but also to take into account that this may also be conditioned by the individual genetic framework. This will have to be characterized by the appropriate technologies (single nucleotide polymorphism, specific allelic combinations, methylation, istilization acetylation, etc.). Although the research status complex on these topics is very promising there is still a lot of work to be done to systematically use biomarkers of the state of nutrition. However, it will still be enough to use existing technologies to standardize and validate a series of biomarker profiles, including biomarkers, in the various aspects of nutrigenomics in order to be able to use this new diagnostic tool. It is also not to be ruled out that from the complex knowledge of the use of nutritional biomarkers, as well as diagnostic applications, they can also gain new opportunities for the development of drugs and therapies related to new knowledge about the pathogenetic processes involved in their function [88].
References
- Ordovas JM, Corella D (2004) Nutritional genomics. Annu Rev Genomics Hum Genet 5: 71-118. [crossref]
- Armstrong B, Doll R (1975) Environmental factors and cancer incidence and mortality in different countries, with special reference to dietary practices. Int J Cancer 15: 617–631.
- Neuhouser ML, Patterson RE, King IB, Horner NK, Lampe JW (2003) Selected nutritional biomarkers predict diet quality. Public Health Nutr 6: 703-709.
- Eilat-Adar S, Goldbourt U (2010) Nutritional recommendations for preventing coronary heart disease in women: evidence concerning whole foods and supplements. Nutr Metab Cardiovasc Dis 20: 459-466.
- Milner JA (2004) Molecular targets for bioactive food components. J Nutr 134: 2492S-2498S. [crossref]
- Lee SA (2009) Gene-diet interaction on cancer risk in epidemiological studies. J Prev Med Public Health 42: 360-370. [crossref]
- Evans WE, Johnson JA (2001) Pharmacogenomics: the inherited basis for interindividual differences in drug response. Annu Rev Genomics Hum Genet 2: 9-39.
- Evans WE, McLeod HL (2003) Pharmacogenomics--drug disposition, drug targets, and side effects. N Engl J Med 348: 538-549. [crossref]
- Brouwer IA, Zock PL, van Amelsvoort LG, Katan MB, Schouten EG (2002) Association between n-3 fatty acid status in blood and electrocardiographic predictors of arrhythmia risk in healthy volunteers. Am J Cardiol 89: 629-631.
- Sacks FM, Katan M (2002) Randomized clinical trials on the effects of dietary fat and carbohydrate on plasma lipoproteins and cardiovascular disease. Am J Med 113: S13-S24.
- Ohlsson L (2010) Dairy products and plasma cholesterol levels. Food Nutr Res 54. [crossref]
- Francis GA, Fayard E, Picard F, Auwerx J (2003) Nuclear receptors and the control of metabolism. Annu Rev Physiol 65: 261-311. [crossref]
- Willett WC (2000) Nutritional epidemiology issues in chronic disease at the turn of the century. Epidemiol Rev 22: 82-86.
- Churchill GA (2002) Fundamentals of experimental design for cDNA microarrays. Nature Genet 32: 490–495.
- Lee CK, Allison DB, Brand J, Weindruch R, Prolla TA (2002) Transcriptional profiles associated with aging and middle age-onset caloric restriction in mouse hearts. Proc Natl Acad Sci USA 99: 14988–14993.
- Cao SX, Dhahbi JM, Mote PL, Spindler SR (2001) Genomic profiling of short- and long-term caloric restriction effects in the liver of aging mice. Proc Natl Acad Sci USA 98: 10630–10635.
- Xiao J, Gregersen S, Kruhøffer M, Pedersen SB, Ørntoft TF, et al. (2001) The effect of chronic exposure to fatty acids on gene expression in clonal insulin-producing cells: studies using high density oligonucleotide microarray. Endocrinology 142: 4777–4784.
- Park JH, Ahn J, Kim S, Kwon DY, Ha YT (2010) Murine hepatic miRNAs expression and regulation of gene expression in diet-induced obese mice. Mol Cells 31: 33-38. [crossref]
- Reed BD, Charos AE, Szekely AM, Weissman SM, Snyder M (2008) Genome-wide occupancy of SREBP1 and its partners NFY and SP1 reveals novel functional roles and combinatorial regulation of distinct classes of genes. PLoS Genet 25: e1000133.
- Grody WW (2003) Molecular genetic risk screening. Annu Rev Med 54: 473-490. [crossref]
- Sachidanandam R, Weissman D, Schmidt SC, Kakol JM, Stein LD, et al. (2001) A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 409: 928-933.
- Blau N, van Spronsen FJ, Levy HL (2010) Phenylketonuria. Lancet 376: 1417-1427. [crossref]
- Terry P, Lichtenstein P, Feychting M, Ahlbom A, Wolk A (2001) Fatty fish consumption and risk of prostate cancer. Lancet 357: 1764-1766. [crossref]
- Morise A, Thomas C, Landrier JF, Besnard P, Hermier D (2009) Hepatic lipid metabolism response to dietary fatty acids is differently modulated by PPARalpha in male and female mice. Eur J Nutr 48: 465-473.
- Dreja T, Jovanovic Z, Rasche A, Kluge R, Herwig R, et al. (2010) Diet-induced gene expression of isolated pancreatic islets from a polygenic mouse model of the metabolic syndrome. Diabetologia 53: 309-320.
- Ness C, Chambers CM (2000) Feedback and Hormonal Regulation of Hepatic 3-Hydroxy-3-Methylglutaryl Coenzyme A Reductase: The Concept of Cholesterol Buffering Capacity Gene. Proc Soc Exp Biol Med 224: 8-19.
- Cheema SK, Clandinin MT (2000) Dietary fat-induced suppression of lipogenic enzymes in B/B rats during the development of diabetes. Lipids 35: 421-425. [crossref]
- Schmitz G, Ecker J (2008) The opposing effects of n-3 and n-6 fatty acids. Prog Lipid Res 47: 147-155. [crossref]
- Chapkin RS, Kim W, Lupton JR, McMurray DN (2009) Dietary docosahexaenoic and eicosapentaenoic acid: emerging mediators of inflammation. Prostaglandins Leukot Essent Fatty Acids 81: 187-91.
- Bouwens M, van de Rest O, Dellschaft N, Bromhaar MG, de Groot LC, et al. (2009) Fish-oil supplementation induces antiinflammatory gene expression profiles in human blood mononuclear cells. Am J Clin Nutr 90: 415-424.
- Bünger M, Hooiveld GJ, Kersten S, Müller M (2007) Exploration of PPAR functions by microarray technology--a paradigm for nutrigenomics. Biochim Biophys Acta 1771: 1046-1064.
- Cobb JP, Mindrinos 2021 Copyright OAT. All rights reservker HV, et al. (2005) Application of genome-wide expression analysis to human health and disease. Proc Natl Acad Sci U S A 102: 4801-4806. [crossref]
- Goyenechea E, Crujeiras AB, Abete I, Martínez JA (2009) Expression of two inflammation-related genes (RIPK3 and RNF216) in mononuclear cells is associated with weight-loss regain in obese subjects. J Nutrigenet Nutrigenomics 2: 78-84.
- Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, et al. (2003) Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest 112: 1796-1808. [crossref]
- Martin KJ, Graner E, Li Y, Price LM, Kritzman BM, et al. (2001) High-sensitivity array analysis of gene expression for the early detection of disseminated breast tumor cells in peripheral blood. Proc Natl Acad Sci U S A 98: 2646-2651.
- Whitney AR, Diehn M, Popper SJ, Alizadeh AA, Boldrick JC, et al. (2003) Individuality and variation in gene expression patterns in human blood. Proc Natl Acad Sci U S A 100: 1896-1901. [crossref]
- Quackenbush J (2006) Computational approaches to analysis of DNA microarray data. Yearb Med Inform . [crossref]
- Lee WN, Go VL (2005) Nutrient-gene interaction: tracer-based metabolomics. J Nutr 135: 3027S-3032S. [crossref]
- Gibney MJ, Walsh M, Brennan L, Roche HM, German B, et al. (2005) Metabolomics in human nutrition: opportunities and challenges. Am J Clin Nutr 82: 497-503.
- Hollywood K, Brison DR, Goodacre R (2006) Metabolomics: current technologies and future trends. Proteomics 6: 4716-4723. [crossref]
- Paolucci U, Vigneau-Callahan KE, Shi H, Matson WR, Kristal BS (2004) Development of biomarkers based on diet-dependent metabolic serotypes: characteristics of component-based models of metabolic serotypes. OMICS 8: 221-238.
- Lindon JC, Holmes E, Nicholson JK (2006) Metabonomics techniques and applications to pharmaceutical research & development. Pharm Res 23: 1075-1088.
- Tsutsui H, Maeda T, Toyo'oka T, Min JZ, Inagaki S, et al. (2010) Practical analytical approach for the identification of biomarker candidates in prediabetic state based upon metabonomic study by ultraperformance liquid chromatography coupled to electrospray ionization time-of-flight mass spectrometry. J Proteome Res 9: 3912-3922.
- Griffin JL (2006) Understanding mouse models of disease through metabolomics. Curr Opin Chem Biol 10: 309-315.
- Gieger C, Geistlinger L, Altmaier E, Hrabé de Angelis M, Kronenberg F, et al. (2008) Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genet 4: e1000282.
- Kotronen A, Velagapudi VR, Yetukuri L, Westerbacka J, Bergholm R, et al. (2009) Serum saturated fatty acids containing triacylglycerols are better markers of insulin resistance than total serum triacylglycerol concentrations. Diabetologia 52: 684-690.
- Walsh MC, Brennan L, Malthouse JP, Roche HM, Gibney MJ (2006) Effect of acute dietary standardization on the urinary, plasma, and salivary metabolomic profiles of healthy humans. Am J Clin Nutr 84: 531e9. [crossref]
- Mao TK, van de Water J, Keen CL, Schmitz HH, Gershwin ME (2002) Modulation of TNF-alpha secretion in peripheral blood mononuclear cells by cocoa flavanols and procyanidins. Dev Immunol 9: 135-141. [crossref]
- Lenz EM, Bright J, Wilson ID, Hughes A, Morrisson J (2004) Metabonomics, dietary influences and cultural differences: a 1H NMR-based study of urine samples obtained from healthy British and Swedish subjects. J Pharm Biomed Anal 36: 841-849. [crossref]
- Lankinen M, Schwab U, Gopalacharyulu PV, Seppänen-Laakso T, Yetukuri L, et al. (2010) Dietary carbohydrate modification alters serum metabolic profiles in individuals with the metabolic syndrome. Nutr Metab Cardiovasc Dis 20: 249-257. [crossref]
- Lankinen M, Schwab U, Erkkilä A, Seppänen-Laakso T, Hannila ML, et al. (2009) Fatty fish intake decreases lipids related to inflammation and insulin signaling--a lipidomics approach. PLoS One 4: e5258. [crossref]
- Miccheli A, Marini F, Capuani G, Miccheli AT, Delfini M, et al. (2009) The influence of a sports drink on the postexercise metabolism of elite athletes as investigated by NMR-based metabolomics. J Am Coll Nutr 28: 553-564. [crossref]
- de Hoog CL, Mann M (2004) Proteomics. Annu Rev Genomics Hum Genet 5: 267-293. [crossref]
- Saleem RA, Rogers RS, Ratushny AV, Dilworth DJ, Shannon PT, et al. (2008) Integrated phosphoproteomics analysis of a signaling network governing nutrient response and peroxisome induction. Mol Cell Proteomics 9: 2076-2088. [crossref]
- Pan S, Chen R, Crispin DA, May D, Stevens T, et al. (2011) Protein Alterations Associated with Pancreatic Cancer and Chronic Pancreatitis Found in Human Plasma using Global Quantitative Proteomics Profiling. J Proteome Res 10: 2359-2376. [crossref]
- Cañas B, López-Ferrer D, Ramos-Fernández A, Camafeita E, Calvo E (2006) Mass spectrometry technologies for proteomics. Brief Funct Genomic Proteomic 4: 295-320. [crossref]
- Kussmann M, Krause L, Siffert W (2010) Nutrigenomics: where are we with genetic and epigenetic markers for disposition and susceptibility? Nutrition Reviews 68: S38-S47. [crossref]
- Kussmann M, Panchaud A, Affolter M (2010) Proteomics in nutrition: status quo and outlook for biomarkers and bioactives. J Proteome Res 9: 4876-4887. [crossref]
- Marvin-Guy L, Lopes LV, Affolter M, Courtet-Compondu MC, Wagnière S, et al. (2005) Proteomics of the rat gut: analysis of the myenteric plexus-longitudinal muscle preparation. Proteomics 5: 2561-2569. [crossref]
- Breikers G, van Breda SG, Bouwman FG, van Herwijnen MH, Renes J, et al. (2006) Potential protein markers for nutritional health effects on colorectal cancer in the mouse as revealed by proteomics analysis. Proteomics 6: 2844-2852. [crossref]
- Tan S, Seow TK, Liang RC, Koh S, Lee CP, et al. (2002) Proteome analysis of butyrate-treated human colon cancer cells (HT-29). Int J Cancer 98: 523-531. [crossref]
- Herzog A, Kindermann B, Döring F, Daniel H, Wenzel U (2004) Pleiotropic molecular effects of the pro-apoptotic dietary constituent flavone in human colon cancer cells identified by protein and mRNA expression profiling. Proteomics 4: 2455-2464. [crossref]
- tom Dieck H1, Döring F, Fuchs D, Roth HP, Daniel H (2005) Transcriptome and proteome analysis identifies the pathways that increase hepatic lipid accumulation in zinc-deficient rats. J Nutr 135: 199-205. [crossref]
- Zhang L, Perdomo G, Kim DH, Qu S, Ringquist S, et al. (2008) Proteomic analysis of fructose-induced fatty liver in hamsters. Metabolism 57: 1115-24. [crossref]
- Davis CD, Milner J (2004) Frontiers in nutrigenomics, proteomics, metabolomics and cancer prevention. Mutat Res 551: 51-64. [crossref]
- Panagiotou G, Nielsen J (2009) Nutritional systems biology: definitions and approaches. Annu Rev Nutr 29: 329-339. [crossref]
- Ideker T, Galitski T, Hood L (2001) A new approach to decoding life: systems biology. Annu Rev Genomics Hum Genet 2: 343-372. [crossref]
- Kitano H (2002) Systems biology: a brief overview. Science 295: 1662-1664. [crossref]
- Desiere F (2004) Towards a systems biology understanding of human health: interplay between genotype, environment and nutrition. Biotechnol Ann Rev 10: 51-84.
- Hucka M, Finney A, Bornstein BJ, Keating SM, Shapiro BE, et al. (2004) Evolving a lingua franca and associated software infrastructure for computational systems biology: the Systems Biology Markup Language (SBML) project. Syst Biol (Stevenage) 1: 41-53. [crossref]
- Le Novère N, Hucka M, Mi H, Moodie S, Schreiber F, et al. (2009) The Systems Biology Graphical Notation. Nat Biotechnol 27: 735-741. [crossref]
- Sauro HM, Hucka M, Finney A, Wellock C, Bolouri H, et al. (2003) Next generation simulation tools: the systems biology workbench and BioSPICE integration. OMICS 7: 355-372. [crossref]
- Funahashi A, Morohashi M, Kitano H, Tanimura N (2003) CellDesigner: a process diagram editor for gene-regulatory and biochemical networks. BIOSILICO 1: 159-162.
- Mendes P, Hoops S, Sahle S, Gauges R, Dada J, et al. (2009) Computational modeling of biochemical networks using COPASI. Methods Mol Biol 500: 17-59.
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW (2010) GenBank. Nucleic Acids Research 37: D26-D31.
- Kouranov A, Xie L, de la Cruz J, Chen L, Westbrook J, et al. (2006) The RCSB PDB information portal for structural genomics. Nucleic Acids Res34: D302-305. [crossref]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, et al. (2010) The Pfam protein families database. Nucleic Acids Res 38: D211-222. [crossref]
- Caspi R, Billington R, Ferrer L, Foerster H, Fulcher CA, et al. (2010) TheMetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res 38: D473-D479. [crossref]
- Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, et al. (2009) Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res 37: D619-D622.
- Kuhn M, Szklarczyk D, Franceschini A, Campillos M, von Mering C, et al. (2010) STITCH 2: an interaction network database for small molecules and proteins. Nucleic Acids Res 38: D552-D556. [crossref]
- Lehne B, Schlitt T (2009) Protein-protein interaction databases: keeping up with growing interactomes. Hum Genomics 3: 291-297. [crossref]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, et al. (2003) Cytoscape: a software Environment for integratedmodels of biomolecular interaction networks. Genome Res 13: 2498-2504. [crossref]
- Maere S, Heymans K, Kuiper M (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448-3449. [crossref]
- Eletto D, Leone A, Bifulco M, Tecce MF (2005) Effect of unsaturated fat intake from Mediterranean diet on rat liver mRNA expression profile: selective modulation of genes involved in lipid metabolism. Nutr Metab Cardiovasc Dis 15: 13-23. [crossref]
- Kaput J (2004) Diet-disease gene interactions. Nutrition 20: 26-31. [crossref]
- Knowles LM, Milner JA (2003) Diallyl disulfide induces ERK phosphorylation and alters gene expression profiles in human colon tumor cells. J Nutr 133: 2901-2906. [crossref]
- Kato S, Fujiki R (2011) Transcriptional controls by nuclear fat-soluble vitamin receptors through chromatin reorganization. Biosci Biotechnol Biochem 75: 410-413. [crossref]
- Rescigno T, Micolucci L, Tecce MF, Capasso A (2017) Bioactive Nutrients and Nutrigenomics in Age-Related Diseases. Molecules 22. [crossref]



